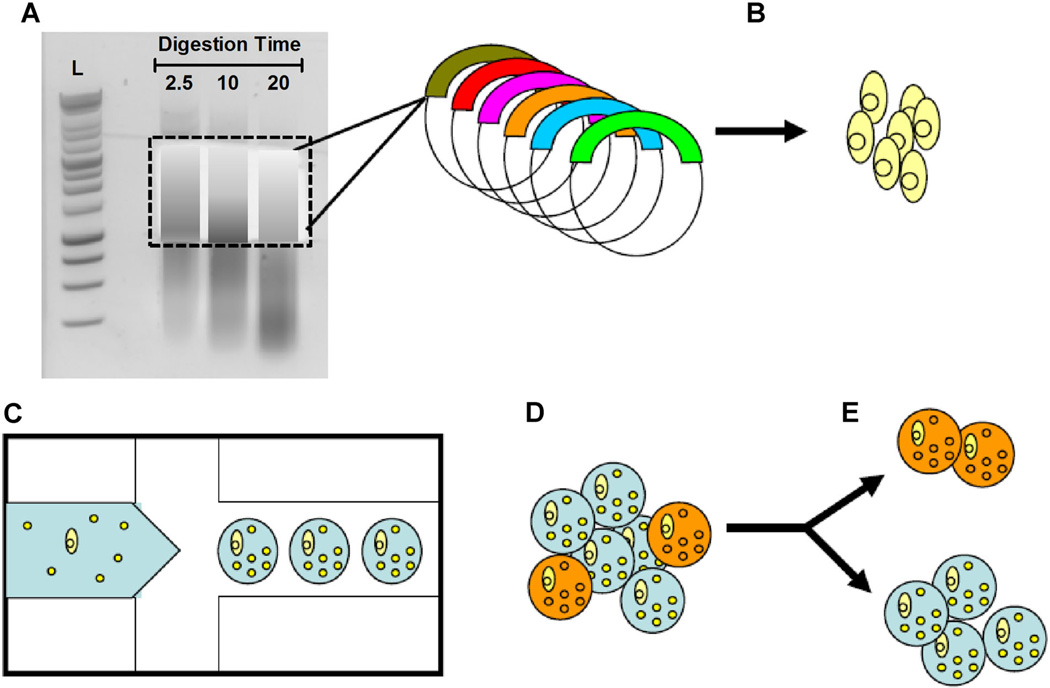
Figure 1.
Schematic of ultra-high-throughput screen for antibiotic drug discovery. A: Environmental DNA was subjected to a limited DNasel digestion and size selected for fragments between 1,000 and 5,000 bp. Digestion time in minutes indicated. B: Metagenomic DNA was cloned into an expression vector by blunt end ligation and then transformed into E. coli bacterial hosts. C: A microfluidic device is used to generate agarose-in-oil micro-emulsions in which individual recombinant E. coli are co-encapsulated with live S. aureus bacterial targets (small yellow spheres). D: GMDs in which the recombinant clone secretes an enzyme that is lytic towards S. aureus are labeled with the SyTox Orange viability probe, which selectively stains only dead bacteria (orange circles). E: Mixed GMD populations are sorted by FACS to isolate E. coli secreting bactericidal natural products. Genes from the sorted GMDs can be cloned, recombined, or rescreened iteratively to facilitate antibiotic drug discovery.