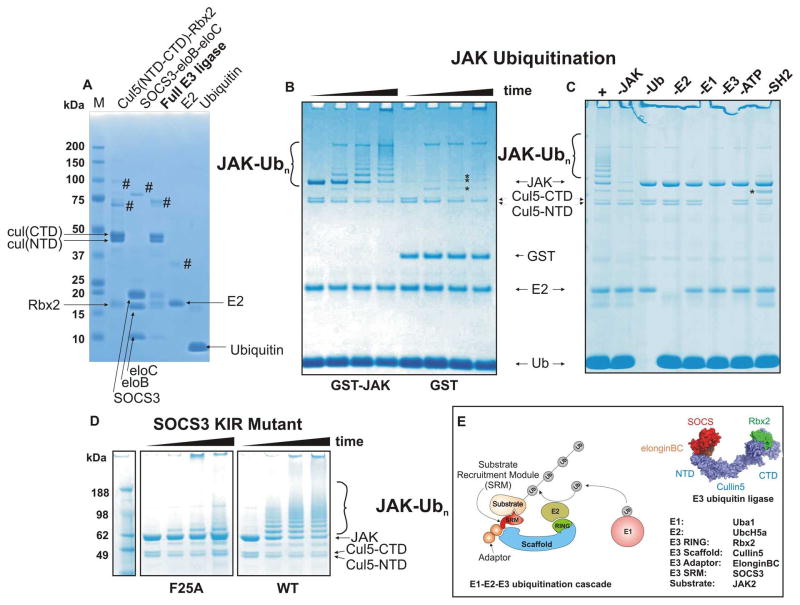
Figure 1. A SOCS3/Cullin5 based E3 ligase catalyses JAK2JH1 ubiquitination.
(A) SDS-PAGE analysis of in vitro ubiquitination system components. Cullin5-Rbx2 and SOCS3-elonginBC are produced separately as ternary complexes (lanes 1,2) and then mixed and further purified to achieve the final E3 ubiquitin ligase. Cullin5 is produced as two domains (NTD and CTD) and these associate non-covalently to form the functional scaffold. Recombinant E2 (UbcH5a) and purified bovine ubiquitin are shown in lanes 4,5 respectively. # marks indicate impurities (B) GST-JAK2 (JH1 domain) is a substrate for polyubiquitination but GST alone is not. SOCS3 based E3 ligase (2.5uM) was incubated with ubiquitin (50uM), human E1 (100nM), purified recombinant E2 (UbcH5a, 2.5uM) and either GST-JAK2 (JH1 domain, 5uM, lanes 1–4) or GST alone (lanes 5–8) in the presence of 2.5mM Mg/ATP at 37°C for 0, 5, 30, 60 minutes. Note that the cullin C-terminal domain was polyubiquitinated in each case (asterisks). (C) GST-JAK polyubiquitination was dependent upon the presence of E1, E2, ATP, ubiquitin and the KIR/SH2 domain of SOCS3. Each reaction component was absent as indicated above the gel. -SH2 indicates that the SH2 domain of SOCS3 was deleted resulting in a SOCS box only:elonginBC ternary complex. (D) Mutating the JAK binding site (F25A) in SOCS3 abrogates its ability to promote JAK ubiquitination The reactions were incubated for 0, 10, 20 and 60 minutes (left to right in each case). All results are visualised by Coomassie staining following SDS-PAGE. (E). Schematic diagram illustrating the components of the Cullin5 in vitro ubiquitination system. A model of the E3 ubiquitin ligase is shown on the right, adapted from PDB 4JGH.