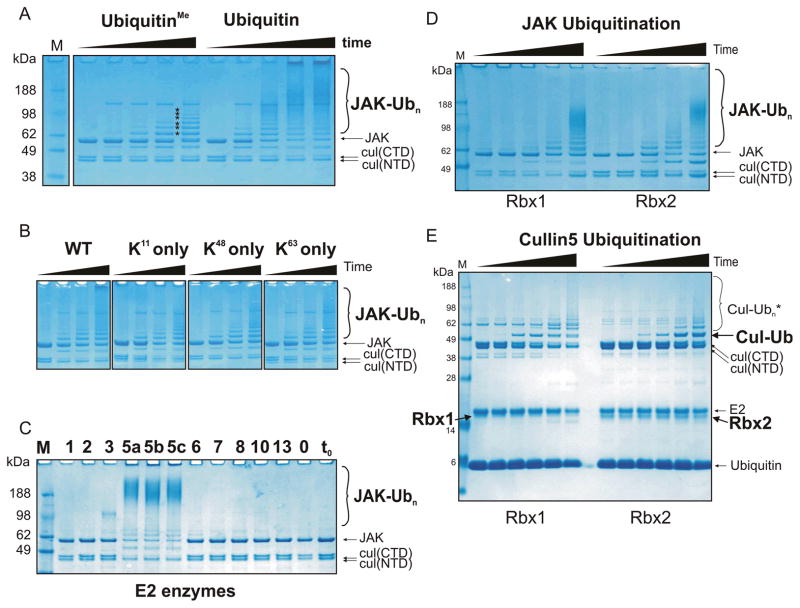
Figure 3. Rbx1 and Rbx2 are active components of a SOCS3/Cullin5 based E3 ligase which catalyses poly-ubiquitination of multiple lysines in the presence of UbcH5a, UbcH5b or UbcH5c.
(A) Ubiquitination reactions were performed as described in Figure 1 using either methylated (left) or wild-type (right) ubiquitin. The use of methylated ubiquitin indicates there are at least six individual lysines on GST-JAK2 (JH1) that are ubiquitinated in this system (asterisks). The reactions were incubated for 0, 10, 30, 60 and 120 minutes (left to right in each case). (B) Polyubiquitination of JAK was observed when ubiquitin with only a single lysine (K11, K48 or K63) was used. (C) A panel of different E2 enzymes were tested for the ability to promote JAK2 ubiquitination. Each E2 enzyme was added to a standard ubiquitination assay (see Figure 1) at a concentration of 2.5 μM and the reaction performed for 30 minutes at 37°C. Only UbcH5a, UbcH5b and UbcH5c were able to promote JAK ubiquitination. The results were visualised by Coomassie staining following SDS-PAGE. (D) Rbx1 and Rbx2 were both active RING finger components of a Cullin5/SOCS3 based E3 ligase. Experiments were performed under identical conditions to those in (A). (E) In the presence of Rbx2, mono-ubiquitinated Cullin5 accumulates to a greater extent than seen in the presence of Rbx1 (indicated by the black label to the right of the gel). Experimental conditions were the same as (D) only JAK was omitted. All results are visualised by Coomassie staining following SDS-PAGE. The ubiquitinated reaction products indicated using larger font at the right of each gel are those referred to in the main text.