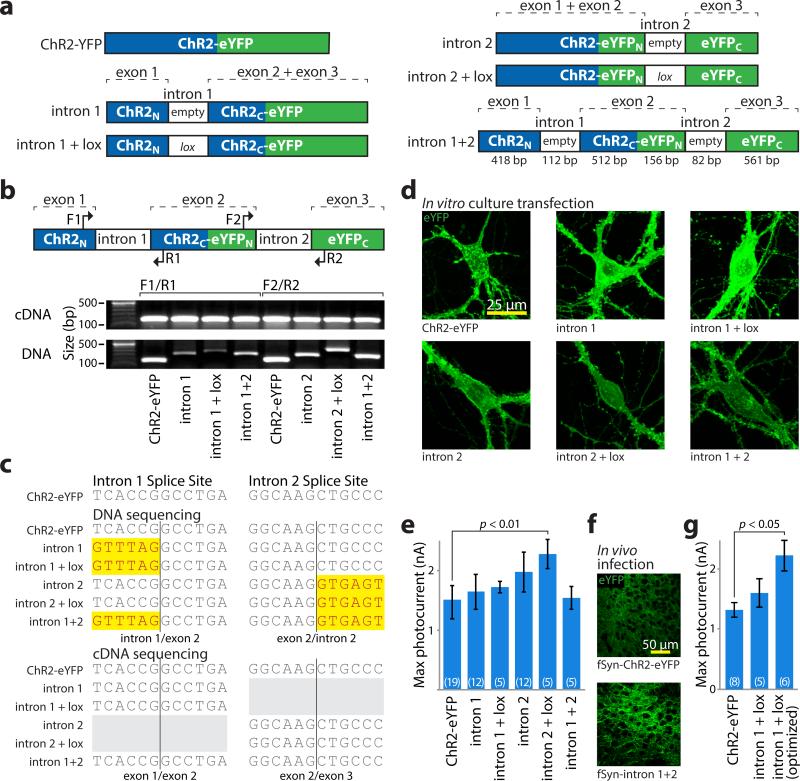
Figure 2. Intron engineering and validation for INTRSECT.
a) Schematic of constructs and nomenclature. ‘empty’ denotes native introns, ‘lox’ indicates addition of lox2722 site (see methods). b) cDNA was prepared from mRNA of HEK293 cells transfected with indicated constructs. Source DNA and cDNA were amplified by PCR with primers (top) in either exons 1 and 2 (‘F1/R1’) or exons 2 and 3 (‘F2/R2’) and separated by gel electrophoresis (bottom). Note that non-spliced DNA bands containing introns vary in size, while cDNA bands from spliced mRNA are uniform. c) Sequence of DNA isolated from bands in b, with uniform removal of intron sequence with these primer sets. d) Images of eYFP expression in primary neuronal cultures transfected with indicated constructs. e) Whole cell photocurrents from primary neurons transfected with intron constructs (ChR2-eYFP: 1422.1±91.7 pA n=19, intron 1: 1590.5±186.7 pA n=12, intron 1 + lox: 1721.7±97.8 pA n=5, intron 2: 1861.6±159.1 pA n=12, intron 2 + lox: 2279.1±246.9 pA n=5, intron 1 + intron 2: 1539.5±188.8 pA n=5, p<0.01 Dunnett's multiple comparison tests ChR2-eYFP vs. intron 2 + lox). All error bars s.e.m. f) Representative images of eYFP expression in medial prefrontal cortex of mouse brain at 2 weeks post-infection timepoint with lentivirus made from indicated constructs (see Supplementary Figure 6 for full dataset). g) Whole cell photocurrents illustrating increased peak photocurrent activity after intron 1 optimization (ChR2-eYFP: 1316±120 pA n=8, intron 1 + lox2722: 1603±236 pA n=5, intron donor: 2219±258 pA n=6, p<0.05 Dunnett's multiple comparison tests, ChR2-eYFP vs. intron donor; see Supplementary Figure 4).