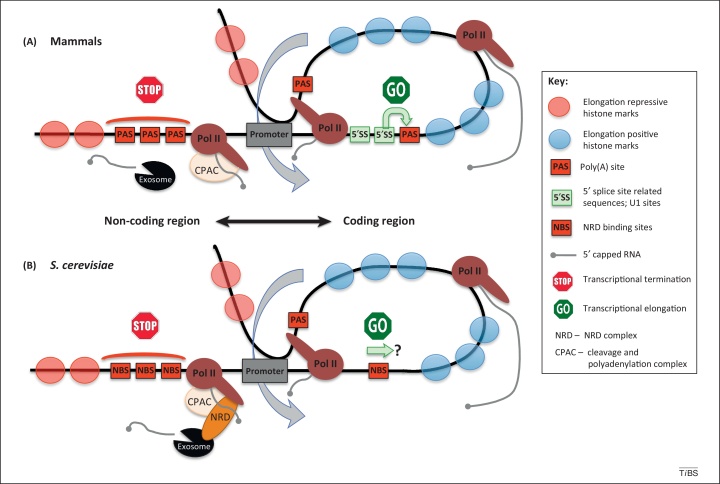
Figure 2.
Mechanisms enforcing promoter directionality. (A) In mammalian cells; (B) in Saccharomyces cerevisiae. Transcription of the upstream noncoding region is restricted by over-representation of termination signals (STOP sign, red rectangles), PAS in mammals or NBS in S. cerevisiae. NBS trigger the NRD termination pathway, which depends on both the NRD complex and CPAC. In contrast PAS-dependent termination is mediated by CPAC only. In both cases released ncRNA is degraded by the exosome. PAS and NBS are depleted over the coding regions. Moreover, in mammalian cells, the coding sequence is enriched in 5′ splice-site-related sequences recognized by U1 small nuclear RNA (snRNA), which inhibits PAS-dependent transcription termination (GO sign). A similar mechanism in yeast is yet to be discovered. Transcriptional directionality is also reinforced by chromatin modification. Nucleosomes with transcription-positive histone marks are presented as blue semitransparent circles, whereas nucleosomes with negative marks are shown in red. A pioneer round of transcription of the coding region establishes a gene loop. Termination factors recruited to Pol II transcribing in the vicinity of the terminator region interacts with initiation factors on the promoter and these juxtapose both regions. The formed loop enhances transcriptional reinitiation into the coding sequence (denoted by bent blue arrows). Abbreviations: CPAC, cleavage and polyadenylation complex; NBS, NRD-binding sites; ncRNA, noncoding RNA; NRD, NRD complex; PAS, poly(A) site; Pol II, RNA polymerase II.