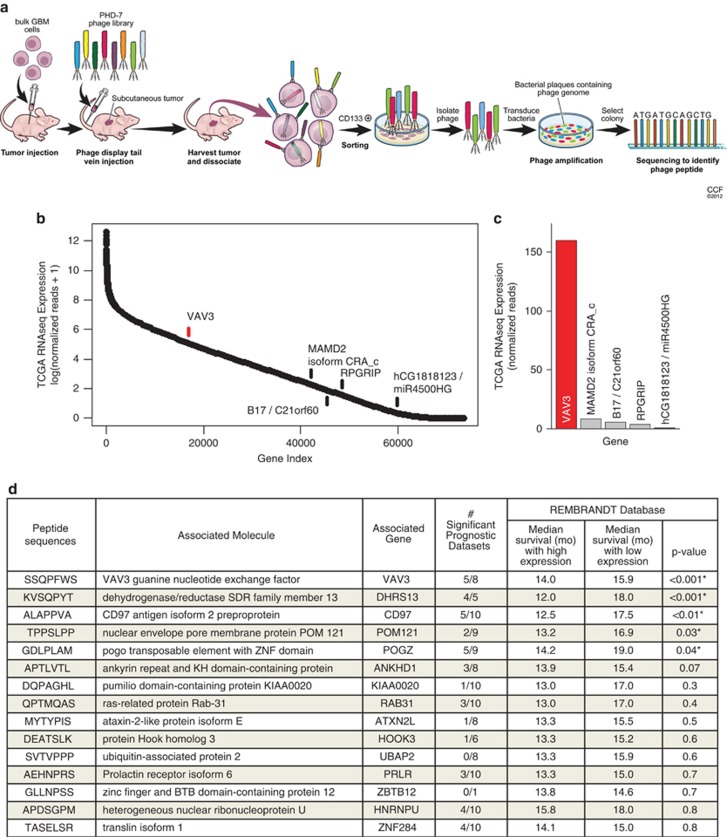
Figure 1.
In vivo phage display identifies GIC-specific peptide sequences. (a) Schematic diagram of in vivo phage display methodology. Freshly dissociated xenografted human glioblastoma cells were subcutaneously implanted into the flank of BALB/c-nu mice. Mice bearing tumors were injected with the phage library through the tail vein. GICs were purified from tumor and bound phages were isolated and sequenced. (b) Peptide sequences obtained using phage display and their corresponding genes were determined by the NCBI BLAST Search function (blast.ncbi.nlm.nih.gov). Analysis of TCGA RNAseq data reveals VAV3 as the most likely phage target for the sequence SSQPFWS. Log scale expression of 73 602 assembled isoform transcripts from TCGA RNA sequencing data were ordered by mean expression of 169 patient tumors with available data. The top scoring sequences by protein BLAST analysis are indicated. (c) Mean number of normalized RNAseq reads, representing expression, of indicated targets identified by protein BLAST is displayed for the sequence SSQPFWS. Both analyses indicate that VAV3 has markedly higher expression in patient glioblastomas than any other potential target. (d) The number of significant prognostic data sets indicates the number of data sets within Oncomine with a statistically significant prognostic survival association with the associated gene (i.e., increased gene expression is associated with significantly decreased survival). Identified genes were then queried in the Rembrandt database to determine the median survival (in months) associated with low versus high expression of the respective gene as well as the statistical significance