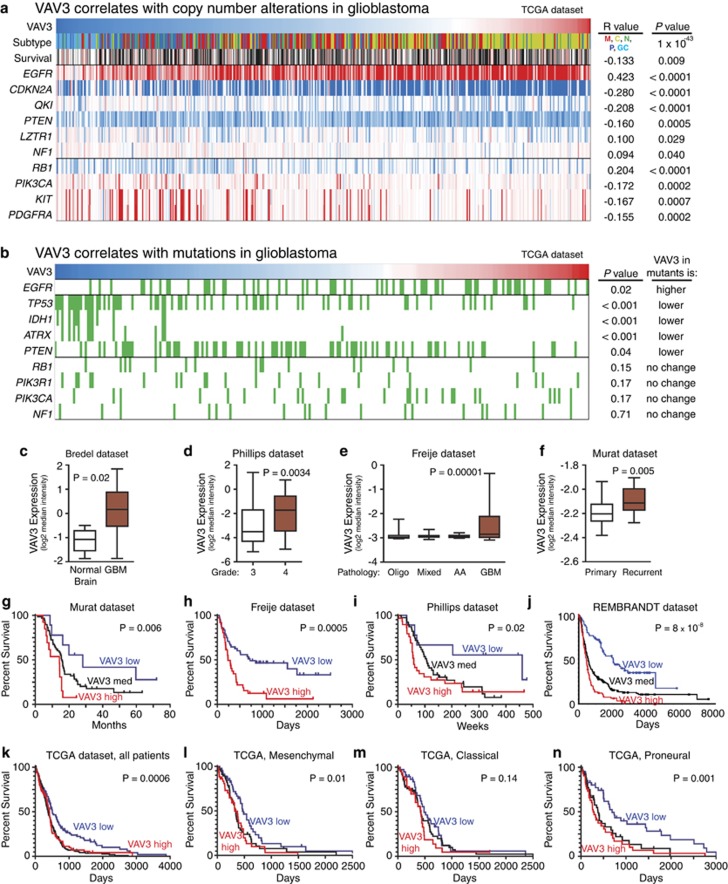
Figure 5.
Bioinformatic analysis reveals VAV3 expression correlates with clinically relevant properties of glioma. (a) Analysis of VAV3 expression and genetic copy number variation. Tumors with high VAV3 expression are more likely to be of the classical subtype, and more likely to have alterations in EGFR, CDKN2A or QKI. VAV3 high tumors are also less likely to have alterations in RB1, PIK3CA, KIT or PDGFRA. Red indicates high expression of VAV3 (top row) or copy number amplification (bottom rows), whereas blue indicates low expression of VAV3 (top row) or copy number loss (bottom rows). (b) Analysis of VAV3 expression and genetic mutation indicates that VAV3 high tumors (red) are more likely to have mutations (indicated in green) in EGFR. Conversely, VAV3 low tumors (blue) are more likely to have mutations in TP53, ATRX, IDH1 and phosphatase and tensin homolog. These results suggest that VAV3 expression is associated with the classical glioblastoma subtype, thereby potentially allowing for more effective anti-VAV3 treatment in a specific subset of patients. (c–f) VAV3 has a statistically significantly greater expression in glioblastoma versus non-malignant brain (c), in glioblastoma versus WHO grade III glioma (d), in glioblastoma versus other brain tumor histology (e) and in recurrent versus primary glioblastoma (f). Among patients with glioblastoma, high expression of VAV3 was associated with significantly shorter survival as compared with low expression as seen in Murat (g), Freije (h), Phillips (i), REMBRANDT (j) and TCGA data sets (k). Within the TCGA subgroups, VAV3 expression correlated with survival in mesenchymal (l) and proneural (n), but not classical (m) patients