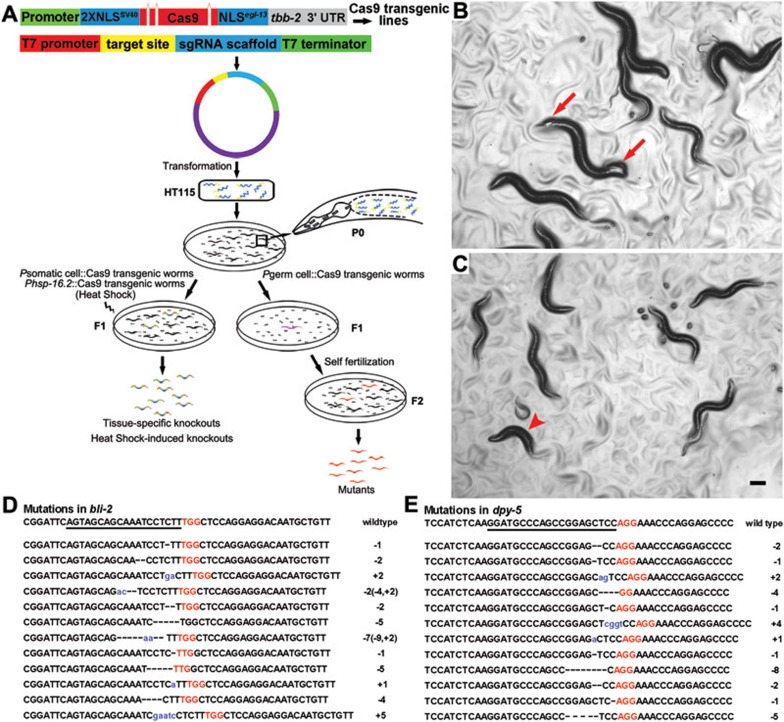
Figure 1.
Inheritable germline mutagenesis and spatial-temporal somatic knockout using the CRISPR-Cas9 feeding system in C. elegans. (A) Schematic description of the CRISPR-Cas9 feeding system. Germline-specific or tissue-specific Cas9 expression vectors were used to generate transgenic lines. Three artificial introns are engineered into the Cas9 coding sequence that is flanked with SV40 and egl-13 NLSs. The resulted cassette is under control of a germline-specific or tissue-specific promoter. To perform the germline or somatic mutagenesis, L4 larval staged Pgerm cell::Cas9 or young adult staged Psomatic cell::Cas9 (P0) worms were fed with HT115 bacteria transformed by the modified pMD18-T vector containing ts-gRNA-46. Individual progeny of P0 (F1) is picked up and seeded onto a culture dish with normal HT115 bacteria. Worms with desired phenotypes could be observed in either F1 (Psomatic cell::Cas9) or F2 (Pgerm cell::Cas9) worms, and are subjected to genotyping or PCR/restriction confirmation. Conditional and direct knockout in somatic lineage of C. elegans can be achieved by feeding transgenic animals, in which the Cas9 gene is under the control of inducible (promoter of heat shock proteins, such as Phsp-16.2) or tissue-specific promoter, with HT115 bacteria containing the plasmid encoding ts-gRNA-46. More details are described in Supplementary information, Figure S1G-S1I, Table S1E-S1F, and Data S1. (B, C) Germline mutagenesis of bli-2 (B) or dpy-5 (C) mediated by the CRISPR-Cas9 feeding system. Worms with strong Bli (arrows) or Dpy (arrowhead) phenotype were observed in F2 worms derived from Ppie-1::Cas9 (P0) fed with (bli-2 or dpy-5)ts-gRNA-46 plasmid-transformed bacteria. Scale bar, 100 μm. (D) Sequences of 12 indels of bli-2 mutants obtained by the CRISPR-Cas9 feeding system. (E) Sequences of 12 indels of dpy-5 mutants obtained by the CRISPR-Cas9 feeding system. The underlined letters are the target sequence (bli-2 or dpy-5). The short lines represent deletions and blue lowercase letters are the insertions. Red capital letters represent the PAM (protospacer adjacent motif), NGG, which must be adjacent to the 3′ end of the target sequence. The nature of the mutations is indicated as the number of base pairs inserted (+) or deleted (−). In cases where both deletion and insertion were observed, details are then explained in parentheses (D).