Abstract
The signalling molecule bis-(3′–5′)-cyclic-dimeric guanosine monophosphate (c-di-GMP) is a central regulator of diverse cellular functions, including motility, biofilm formation, cell cycle progression and virulence, in bacteria. Multiple diguanylate cyclase and phosphodiesterase-domain-containing proteins (GGDEF and EAL/HD-GYP, respectively) modulate the levels of the second messenger c-di-GMP to transmit signals and obtain such specific cellular responses. In the genus Bordetella this c-di-GMP network is poorly studied. In this work, we evaluated the expression of two phenotypes in Bordetella bronchiseptica regulated by c-di-GMP, biofilm formation and motility, under the influence of ectopic expression of Pseudomonas aeruginosa proteins with EAL or GGDEF domains that regulates the c-di-GMP level. In agreement with previous reports for other bacteria, we observed that B. bronchiseptica is able to form biofilm and reduce its motility only when GGDEF domain protein is expressed. Moreover we identify a GGDEF domain protein (BB3576) with diguanylate cyclase activity that participates in motility and biofilm regulation in B. bronchiseptica. These results demonstrate for the first time, to our knowledge, the presence of c-di-GMP regulatory signalling in B. bronchiseptica.
Introduction
Bis-(3′–5′)-cyclic-dimeric guanosine monophosphate (c-di-GMP) is a bacterial second messenger known to regulate a variety of cellular processes. c-di-GMP has been found to stimulate biofilm formation, inhibit motility and control the virulence of bacterial pathogens (Jenal & Malone, 2006; Kolter & Greenberg, 2006; Römling & Amikam, 2006; Hengge, 2009).
c-di-GMP is produced from two molecules of GTP by diguanylate cyclases (DGCs) and hydrolysed by a c-di-GMP-specific phosphodiesterases (PDE). DGC activity is conferred by the GGDEF functional domain, whereas PDE activity is performed by unrelated EAL or HD-GYP domains (Römling & Amikam, 2006). Notably, individual bacterial genomes frequently encode numerous GGDEF and EAL/HD-GYP proteins (Galperin, 2004, 2005), implying that the c-di-GMP network is a highly complex and tightly regulated intracellular signalling system. The observation that most GGDEF and EAL domains are linked directly or through a two-component phosphorylation cascade (Galperin, 2006) to input signal domains (including PAS, blue-light-sensing, Cache, CHASE and MASE domains) implies that numerous environmental and internal signals can be integrated into the c-di-GMP network.
Bordetella bronchiseptica is a Gram-negative bacterium that causes respiratory tract infections in mammals, producing atrophic rhinitis in pigs, kennel cough in dogs and snuffles in rabbits (Goodnow, 1980). B. bronchiseptica has a variety of virulence factors and strategies that allows the host infection. Each factor such as pertactin (PRN), filamentous haemagglutinin (FHA), adenylate cyclase (AC), the type three secretory system (TTSS) and LPS are likely to perform specific functions required for successful colonization (Harvill et al., 1999, 2000; Sisti et al., 2002; Skinner et al., 2004; Inatsuka et al., 2010). The BvgAS two-component system plays a central role in the regulation of some of these factors such as PRN, FHA, AC and TTSS (Mattoo & Cherry, 2005). This system comprises a histidine kinase sensor protein, BvgS, and a DNA-binding response regulator protein, BvgA. In response to environmental signals, BvgAS undergoes a series of phosphorelay signal transduction events that ultimately lead to differential transcription of target genes (Mattoo & Cherry, 2005). Growth at 37 °C in low concentrations of nicotinic acid (NA) and magnesium sulfate leads to the Bvgplus or virulent phase, in which BvgAS activates expression of most known virulence factors. When the Bordetellae are grown at low temperatures (below 26 °C) or in the presence of millimolar concentrations of NA or magnesium sulfate, BvgAS is inactivated, resulting in the Bvgminus or avirulent phase, in which transcription of most virulence factor genes is repressed and the expression of other factors is induced, resulting in the maximal expression of motility loci and genes required for the production of urease (Akerley & Miller, 1993; Akerley et al., 1992; McMillan et al., 1996). Intermediate phase, called Bvgi (Bvg intermediate), occurs transiently during shifts between the Bvgplus and Bvgminus states and can be induced by the addition of magnesium sulfate or NA at concentrations lower than those needed to fully induce the Bvgminus phase (Cotter and Miller, 1997). The Bvgi phase is characterized by the expression of a subset of the Bvgplus-phase genes (such as fhaB and prn) and Bvgi-specific genes (such as bipA and bcfA) (Cotter and Miller, 1997; Stockbauer et al., 2001).
The ability of this genus to form biofilms has been reported in previous work, with other authors reporting the ability of B. bronchiseptica to form biofilm-like structures on abiotic surfaces regulated by the two-component system BvgAS (Irie et al., 2004; Mishra et al., 2005). Static growth with intermediate NA concentrations represented the best conditions for biofilm formation (Mishra et al., 2005). Like in other biofilms, extracellular DNA and exopolysaccharide are important for biofilm formation in B. bronchiseptica (Conover et al., 2011). Sloan and colleagues even observed these structures in vivo in the nasal epithelium of mice infected with B. bronchiseptica. Those structures present a polysaccharide essential for in vivo biofilm development (Sloan et al., 2007). However, the exact mechanism of this process and the bacterial factors involved has not been yet determined.
Although the cyclic-di-GMP signalling system has been characterized in several types of bacteria regulating biofilm formation and motility, the understanding of this important signalling mechanism is limited in Bordetella spp. So far there are no reports relating the c-di-GMP network to a specific phenotype in B. bronchiseptica. In the published genome of B. bronchiseptica RB50 strain there are four hypothetical proteins with EAL domains, ten with GGDEF domains and five with both domains (Amikam & Galperin, 2006). A recent study has identified a GGDEF diguanylate cyclase type protein with ability to synthesize c-di-GMP from the genome sequence of B. pertussis Tohama I. Deletion of the gene encoding this enzyme diminished the capacity of B. pertussis to adhere to abiotic surfaces (Wan et al., 2009).
In the present work, we demonstrated that c-di-GMP signalling is present in B. bronchiseptica by introducing genes coding for proteins with proven PDE or DGC activity. Phenotypes previously linked with the presence of c-di-GMP in other bacteria were analysed, particularly biofilm formation and motility in soft agar. We also describe BB3576, a probable DGC, involved in swimming motility regulation and biofilm formation. The results reported here strongly suggest that c-di-GMP regulates these phenotypes in B. bronchiseptica.
Methods
Bacterial strains and growth conditions.
Strains and plasmids used in this study are listed in Table S1 available with the online version of this paper. B. bronchiseptica strains were grown on Bordet Gengou agar (Britania) supplemented with 10 % (v/v) defibrinated fresh sheep blood (BGA medium) at 37 °C for 48 h and replated in the same medium for 24 h. Escherichia coli (DH5-α and S17-1) and Pseudomonas fluorescens Pf0-1 strains were routinely cultured with liquid LB medium in a test tube or on solidified LB with 1.5 % (w/v) agar. Proteins with DGC (PA1120) and PDE (PA3947) domains were overexpressed in B. bronchiseptica 9.73 by using vectors with the inducible tac promoter to assess the consequences of alterations in c-di-GMP levels. Plasmids were introduced into B. bronchiseptica or in Pseudomonas fluorescens Pf0-1 by electroporation and transformants were selected in the presence of gentamicin (50 µg ml−1).
B. bronchiseptica protein BB3576 was overexpressed in B. bronchiseptica by using broad-host-range plasmid pBBR1MCS-5 with a constitutive promoter nptII (Dombrecht et al., 2001) cloned by us in SacI and XbaI sites of the pBBR1MCS-5 multiple cloning site. Proof reading Pfx taq polymerase was employed to amplify the BB3576 gene from B. bronchiseptica 9.73 with primers BB3576FXhoI (5′ATGCCTCGAGGACGGGGTCGGATAAGGATA3′) and BB3576RKpnI (5′CTGAGGTACCTTCGGTCAGGAACGCAGGTC3′). PCR conditions were 94 °C for 4 min (1 cycle), followed by 35 cycles of 94 °C for 20 s, 58 °C for 20 s, and 68 °C for 2 min and a final extension of 68 °C for 5 min. Underlined portions indicated restriction enzyme sites. Underlined portions indicated restriction enzyme sites. The PCR product was digested with XhoI and KpnI and cloned into pBB1RMCS-5-nptII (renamed pEmpty) that had been digested with the same enzymes, to generate pBB3576. Constructs were confirmed by sequencing. Plasmids were transferred into B. bronchiseptica 9.73 or into Pseudomonas fluorescens Pf0-1 by electroporation and transformants were selected in the presence of gentamycin (50 µg ml−1).
Biofilm assays.
B. bronchiseptica biofilm formation assays using static cultures were performed as described previously (Irie et al., 2004) from overnight cultures into Stainer–Scholte (SS) liquid medium (Stainer & Scholte, 1970). The overnight culture was diluted to OD650 0.1, pipetted into each well of a sterile 96-well U-bottom microtitre plate (both polycarbonate and PVC surfaces) and incubated statically at 37 °C with NA or magnesium sulfate at different concentrations. 1 mM IPTG and gentamicin (50 µg ml−1) were added, when appropriate.
For P. fluorescens biofilms an aliquot (1.5 µl) of an overnight culture grown in LB was transferred into 100 µl K10T-1 medium in a 96-well plate (353911; BD Falcon) and grown statically for 6 h at 30 °C. K10T-1 medium, used in prior studies as a biofilm-promoting, phosphate-rich medium, contains 50 mM Tris/HCl (pH 7.4), 0.2 % (w/v) Bacto tryptone, 0.15 % (v/v) glycerol, 0.6 mM MgSO4 and 1 mM K2HPO4 and was prepared as described previously (Newell et al., 2011).
In all assays planktonic bacteria were removed and remnant cells were stained with 0.1 % crystal violet solution (CV). The stain was dissolved by adding 120 µl 33 % acetic acid solution. A 100 µl sample was transferred to a microplate and then quantified by measuring OD595. Each data point results from an average for six wells and each experiment was repeated at least three times with similar results. Figures are representative of one of those replicates.
Measurements of c-di-GMP levels.
c-di-GMP levels were analysed via LC-MS. Strains of interest were grown in BGA media for 24 h. Four replicates of each strain was harvested and resuspended in 250 µl extraction buffer [methanol : acetonitrile : water (40 : 40 : 20) plus 0.1 M formic acid at −20 °C] and incubated at −20 °C for 30 min. The cell debris was pelleted for 5 min at 4 °C, and the supernatant containing the nucleotide extract was saved. Samples were immediately adjusted to a pH of approximately 7.5 with 15 % (NH4)2HCO3 and stored on dry ice prior to analysis. The resultant extract was analysed via LC-MS using the LC-20AD HPLC system (Shimadzu) coupled to a Finnigan TSQ Quantum Discovery MAX triple-quadrupole mass spectrometer (Thermo Electron) as previously described (Newell et al., 2011).
Motility assays.
SS soft-agar motility plate [0.35 % (p/v) agar] supplemented with MgSO4 40 mM and 1 mM IPTG was used to determine the motility of bacterial strains as previously described (Fernández et al., 2005). The diameter of the migration zone was measured after 18 h of incubation at 37 °C.
Scanning electron microscopy (SEM).
Bordetella strains were cultured statically on glass coverslips partially submerged vertically in 1.5 ml plastic tubes (Mishra et al., 2005). The glass coverslips were placed such that an air–liquid interface was established. After 48 h the coverslip was removed and washed with sterile PBS (KH2PO4 3 mM, Na2HPO4 10 mM, NaCl 120 mM), and the bacteria were fixed with 2.5 % glutaraldehyde. Samples were dehydrated in a graded ethanol series (20, 50, 70, 90 and 100 % for 60 min each), dried by critical point using liquid carbon dioxide (EMITECH, K850) and sputter coated with gold. The surface topographies of the biofilm were visualized with a scanning electron microscope (Philips SEM 505), and the images were processed [Image Soft Imaging System ADDA II (SIS)].
RNA extraction and qPCR quantification.
RNA preparation from bacteria was performed using the Illustra RNAspin kit (GE). Samples were placed on ice, and quantification of RNA was performed using a ND-1,000 NanoDrop spectrophotometer at 260 nm. Measurements of A260/A280 were used to determine the purity of the RNA. DNase (Promega) treatment was performed according to manufacturer’s instructions in order to eliminate contaminating DNA. The synthesis of cDNA was performed with a Reverse Transcription System kit (Promega) according to the manufacturer’s protocol using random primers. For each sample 1 µg RNA was used. The reaction was incubated at room temperature for 10 min, and reverse transcription was performed in a thermal cycler at 42 °C for 15 min and 95 °C for 5 min. qPCR conditions were 95 °C for 10 min (1 cycle), followed by 40 cycles of 60 °C for 30s, 95 °C for 15 s and a final increasing temperature cycle between 55 and 95 °C for 10 s when melt curve data was obtained. Samples were placed on ice for 5 min to stop the reaction. Samples that had not undergone the reverse transcription process were used as controls for genomic DNA absence in real-time PCR experiments. Only samples with threshold cycle (Ct) values above mock values were used in further experiments. Real-time PCR were performed using the SYBR green master mix 2× (Bio-Rad). Primers employed are summarized in Table S2. All samples, including a negative control using mock samples instead of DNA, were run in triplicates. The data were analysed using Ct calculations using recA expression levels as a normalizer. Results were expressed as fold increase over values from bacteria growth in SS media without additional NA.
Statistical analyses.
All the results were compared by ANOVA followed by the Tukey test. A value of P<0.01 was considered significant. To identify significant differences in RNA expression, Ct values were analysed using the REST program.
Results
B. bronchiseptica biofilm formation is enhanced by diguanylate cyclase activity
Because some authors have described that not all proteins with GGDEF domains have DGC activity, we first decided to use a protein with known in vitro and in vivo DGC activity in our studies (Kulesekara et al., 2006). Protein PA1120 with DGC activity was overexpressed in B. bronchiseptica 9.73 (Bb-DGC) by using a vector with the inducible tac promoter. Clone Bb-DGC grown in IPTG-containing media overexpressed PA1120 and this strain was able to grow in a batch culture at the same rate as the parental strain (data not shown). The ability to form biofilm was evaluated by the crystal violet method in 96-well polycarbonate U-bottom and polyvinylchloride (PVC) microtitre plates as described in Methods. Biofilm formation is regulated by the BvgAS two-component system (Irie et al., 2004; Mishra et al., 2005). Hence, we determined the amount of biofilm biomass formed by strains under Bvg-modulated-phase conditions in the presence of NA (0–8.0 mM). This chemical compound serves as an environmental modulator of BvgAS activity (Akerley et al., 1992), at 0 mM NA the BvgAS system should be fully activated or in Bvgplus phase and at 8 mM NA the BvgAS system should be fully inactivated or in Bvgminus phase. In our hands, B. bronchiseptica 9.73 (BbWT) strain showed biofilm formation on both polycarbonate and PVC surfaces and the biofilm pellicle was enhanced in intermediate modulation conditions in agreement with results for B. bronchiseptica RB50 strain (Irie et al., 2004). In Fig. 1(a) results obtained on polycarbonate surface are shown and are similar to PVC results (not shown). As expected for a high c-di-GMP intracellular concentration, overexpression of PA1120 in Bb-DGC significantly enhanced pellicle formation at NA concentrations from 0 to 4.0 mM (Fig. 1a) compared with the parental strain. This difference was observed only when IPTG-supplemented SS was used. Surprisingly, when PA1120 was overexpressed biofilm formation was particularly exacerbated in the Bvgminus phase (4.0 mM NA; Fig. 1a). We observed no biofilm formation in 8.0 mM NA media, but planktonic growth in that condition was significantly slower compared with that for media without additional NA (data not shown). On the other hand, overexpression of PA3947 with PDE activity produced a biologically significant decrease in the biofilm formation as compared with the parental strain only in 1.0 mM NA (Fig. 1a).
Fig. 1.
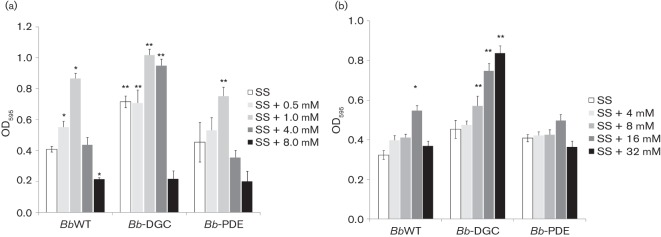
Effect of DGC or PDE overexpression in B. bronchiseptica biofilm formation. Biofilm development by BbWT, Bb-DGC and Bb-PDE cultured in SS medium either alone or supplemented with NA (a) or magnesium sulfate (b). Biofilm formation was assessed by polycarbonate microtitre plate assays as described in Methods. * P<0.01 vs BbWT grown in SS; ** P<0.01 vs BbWT at the same modulator concentration.
The BvgAS system is downregulated by NA and by magnesium sulfate. For that reason, we performed biofilm quantification with different magnesium sulfate concentrations corresponding to Bvgplus, Bvgi and Bvgminus phases, as reported previously (Williams & Cotter, 2007). We observed that biofilm formation was also enhanced in Bb-DGC at Bvgi (16 mM) and Bvgminus (32 mM) magnesium sulfate-induced phases compared with the parental strain (Fig. 1b).
In order to confirm that NA concentrations corresponded to established phases in BbWT like in the sequenced B. bronchiseptica strain RB50, mRNA extraction and quantification was performed. Bacteria cultures grown in SS with NA at concentrations between 0 and 4.0 mM were subject to mRNA purification. Levels of cyaA (AC, Bvgplus phase factor), bipA (Bvgi phase factor) and flaA (flagellin, Bvgminus phase factor) mRNA were normalized to recA and compared. Results obtained confirmed that with NA in the concentrations assayed all the virulence phases were detected, 0.5 mM NA and 1 mM NA correspond to Bvgi phase and 4 mM NA corresponds to Bvgminus phase. (Fig. 2).
Fig. 2.
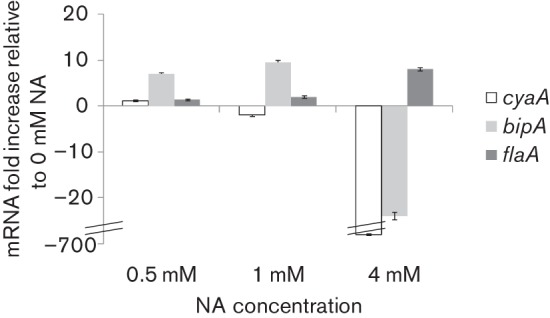
mRNA expression of B. bronchiseptica 9.73 Bvg-regulated factors at different nicotinic acid concentrations. mRNA transcripts levels of cyaA (Bvgplus gene), bipA (Bvgi gene) and flaA (Bvgminus gene) were quantified by qPCR compared with the constitutive recA gene. Values were normalized to levels observed in SS media without additional NA (Bvgplus phase).
Overexpression of a diguanylate cyclase in B. bronchiseptica induced increased intracellular c-di-GMP levels
In order to confirm that PA1120 overexpression in Bb-DGC correlated with increased DGC activity in vivo, intracellular c-di-GMP levels were determined. Cultures with or without IPTG induction were subjected to c-di-GMP extraction and analysis. As shown in Fig. 3, Bb-DGC in the presence of IPTG presented significantly higher c-di-GMP levels than the strain grown under non-inducing conditions, supporting the hypothesis that biofilm phenotypes observed in Bb-DGC were a consequence of increased c-di-GMP level.
Fig. 3.
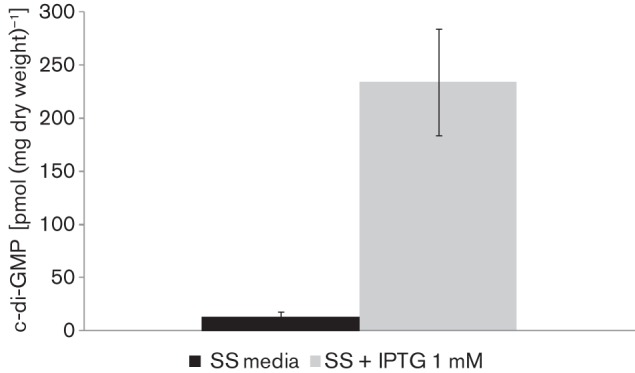
In vivo assessments of DGC activity in B. bronchiseptica. Quantitative measurements of cellular c-di-GMP from the Bb-DGC strain compared with BbWT. Formic acid-extracted c-di-GMP was measured by LC-MS and normalized to mg dry weight of bacteria after extraction. The difference observed was significant (P<0.001).
Scanning electron microscopy of B. bronchiseptica cells adhered to glass coverslips
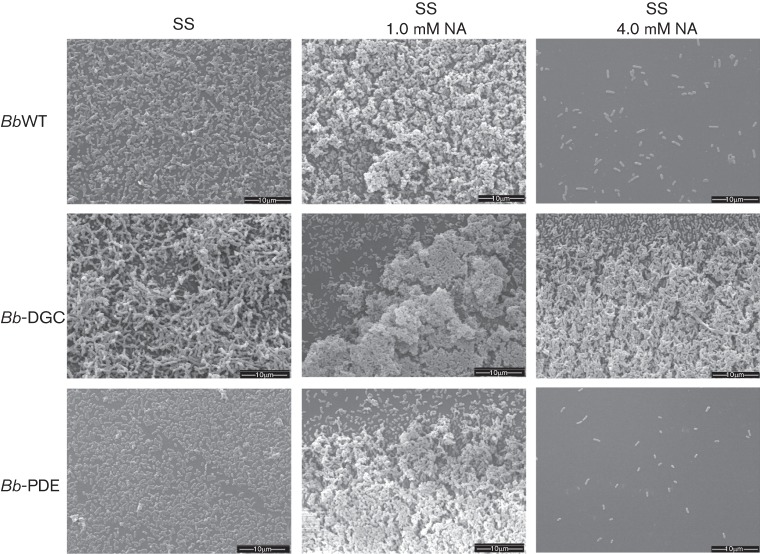
To further investigate the consequences of DGC activity in biofilm development, biofilms formed at the air–liquid interface in biphasic cultures on glass coverslips were observed by SEM. As expected for Bvgi conditions, at 48 h all strains grown in 1.0 mM NA presented thick multilayered stack of cells alternated with stacked cells aggregated in clusters similar to those observed by other authors (Fig. 4) (Mishra et al., 2005). Wild-type cells grown in Bvgplus phase (0 mM NA) formed a thin layer with small aggregates, whereas cells grown in Bvgminus phase (4.0 mM NA) appear as diffuse, interspersed bacteria (Fig. 4). Interestingly, SEM analysis of Bb-DGC biofilms revealed stacked bacteria more frequently and with bigger dimensions, resulting in an architecture that appeared to encase the bacterial microcolonies. These structures were observed in all virulence phases tested. In the case of Bb-PDE, cells grown at Bvgplus (0 mM NA) or Bvgminus (4.0 mM NA) phase were spread around the disks with large regions remaining uncolonized. Altogether these results further corroborate those from CV quantification for Bb-DGC, with the exception that no differences were observed at 1.0 mM NA between Bb-PDE and BbWT in this analysis.
Fig. 4.
SEM images of B. bronchiseptica biofilms. BbWT (upper row of panels), Bb-DGC (middle row of panels) and Bb-PDE (bottom row of panels) were grown statically on vertically submerged coverslips in SS medium alone (left column of panels), SS supplemented with 1.0 mM NA (middle column of panels) or SS supplemented with 4.0 mM NA (right column of panels). After 48 h of growth, the biofilms formed at the air–liquid interface were visualized by SEM.
BvgA is not necessary for diguanylate-cyclase-dependent biofilm formation
Bacterial proteins that mediate c-di-GMP turnover and signal transduction are often composed of multiple domains, allowing for a variety of regulatory inputs (Galperin et al., 2001). In particular, interplay between c-di-GMP network and two-component systems has been observed in many bacteria (Lai et al., 2009; Mikkelsen et al., 2009). B. bronchiseptica biofilm formation was dependent on Bvg phase conditions and c-di-GMP signalling as described above, indicating a probable connection between the two regulatory systems.
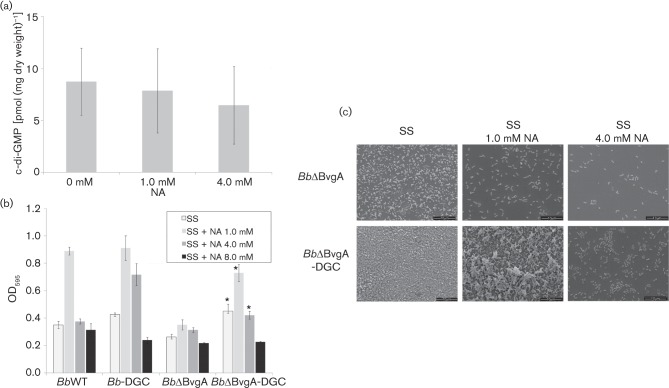
As described above, biofilm formation was dependent on Bvg phase condition even if high c-di-GMP intracellular levels were induced. A hypothesis to explain this observation is that the BvgAS system controls c-di-GMP levels. However, quantification of intracellular c-di-GMP levels under different Bvg-modulated conditions shows no significant differences between c-di-GMP concentrations (Fig. 5a), thus we must reject this hypothesis.
Fig. 5.
BvgA is not necessary for diguanylate-cyclase-dependent biofilm formation. (a). Quantitative measurements of cellular c-di-GMP from B. bronchiseptica grown planktonically in BGA supplemented with different NA concentrations. Formic-acid-extracted c-di-GMP was measured by LC-MS and normalized to mg dry weight of bacteria after extraction. (b) Biofilm formation by BbWT, Bb-DGC, BbΔBvgA and BbΔBvgA-DGC cultured in SS medium either alone or supplemented with NA was analysed by CV staining after 24 h in static conditions. (c) SEM of 48 h air–liquid interface submerged coverslips in SS medium alone (left panels), SS supplemented with 1.0 mM NA (middle panels) or SS supplemented with 4.0 mM NA (right panels). * P<0.01 vs BbΔBvgA grown at the same NA concentration.
Although c-di-GMP concentration was apparently independent of direct BvgAS regulation, we assessed the effect of a bvgA mutant in a strain overexpressing a DGC. We overexpressed PA1120 with DGC activity in B. bronchiseptica ΔBvgA (BbΔBvgA), a mutant locked in the BvgAminus phase and previously constructed by our group (Fernández et al., 2005). All strains were assayed for biofilm formation in 96-well polycarbonate U bottom plates. As expected, BbΔBvgA showed poor biofilm formation in all Bvg modulatory conditions, similar to the BvgAminus phase (Fig. 5b). However, when PA1120 in BbΔBvgA-DGC was overexpressed, significantly higher biofilm formation was detected at NA concentrations lower than 8.0 mM.
Biofilms produced by DGC-expressing bacteria in a ΔBvgA background were analysed by SEM. As shown in Fig. 5(c), BbΔBvgA-DGC in 1.0 mM NA was the only strain and condition that formed a uniform multilayered stack of cells, but the volume of the structure was smaller than that for Bb-DGC, corroborating the CV determinations (Fig. 4). As expected, BbΔBvgA showed no distinct region of biofilm formation and cells were spread evenly across the coverslips (Fig. 5c).
c-di-GMP regulates swimming motility in B. bronchiseptica
In several bacteria c-di-GMP suppresses swimming, swarming and twitching motility (Jenal & Malone, 2006; Römling & Amikam, 2006). Flagellar expression and motility in B. bronchiseptica is regulated by the BvgAS two-component system and is maximal in the BvgAminus phase (Akerley et al., 1992). Therefore, we evaluated strain motility in the presence of modulating concentrations of magnesium sulfate. As expected, all strains showed motility in soft agar in modulated conditions. However, when DGC overexpression was induced by IPTG in WT reduced motility was observed (Fig. 6). This phenotype was observed also in a ΔBvgA background. Bb-PDE showed no significant differences from the wild-type strain with regard to motility.
Fig. 6.
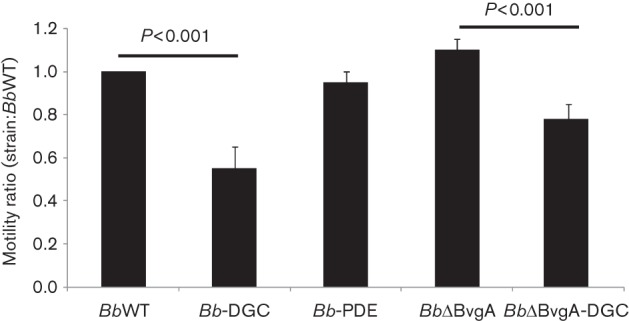
Motility phenotypes of B. bronchiseptica strains. The diameters of migration zones of the wild-type, overexpression strains and mutants were measured and expressed relative to wild-type diameter after 18 h of incubation at 37 °C on SS motility plates supplemented with 32 mM magnesium sulfate. One hundred per cent motility in BbWT was 12 mm. The results are based on three replicates.
BB3576: a hypothetical GGDEF protein regulating swimming motility and biofilm formation in B. bronchiseptica
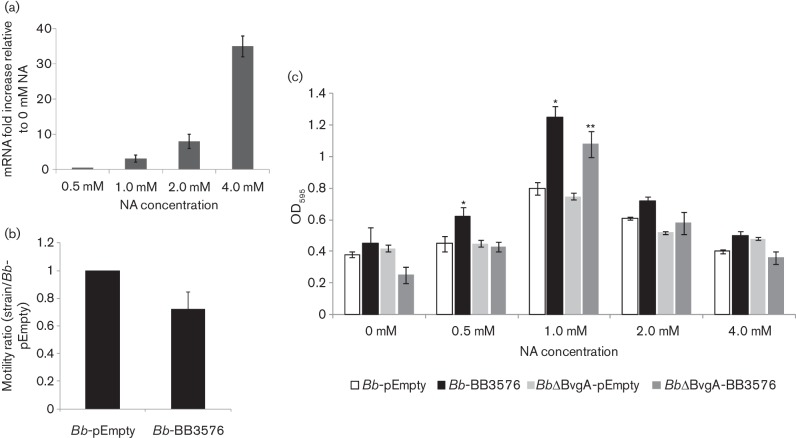
The B. bronchiseptica genome contains 15 proteins with GGDEF domains. As observed above, DGC overexpression in B. bronchiseptica inhibits swimming motility in soft agar. In E. coli, swimming is regulated by c-di-GMP through diverse DGCs and PDEs. Deletion of genes encoding GGDEF proteins YeaI, YedQ, YfiN, YeaJ and YneF in E. coli increased swimming motility (Sanchez-Torres et al., 2011). Also, YeaJ, YedQ and YfiN overexpression negatively regulate motility without impairing it completely (Pesavento et al., 2008). These results suggested that some proteins with DGC activity are involved in fine-tuning regulation of flagellar activity. Among these proteins, YeaJ also has a Cache domain similar to the domain found in the BB3576 protein of B. bronchiseptica. Swimming motility in B. bronchiseptica is regulated mainly by the two-component system BvgAS and is fully expressed in Bvgminus phase. Hence we hypothesized that if BB3576 is involved in motility regulation it should be expressed in the Bvgminus phase. Bacteria grown in the presence of NA were subjected to RNA extraction and subsequent cDNA synthesis. qPCR relative quantification of BB3576 mRNA demonstrated that BB3576 is expressed in all phases tested and 36 times higher in the Bvgminus than in the Bvgplus phase, suggesting that BB3576 protein may be present when flagellar apparatus is functionally active (Fig. 7a).
Fig. 7.
Motility and biofilm phenotypes of B. bronchiseptica overexpressing BB3576. (a) mRNA transcripts levels of BB3576 were quantified by qPCR compared with the constitutive recA gene. Values were normalized to levels observed in SS media without additional NA. (b) The diameters of migration zones of the wild-type strain with pBBR1MCS-5 plasmid alone (Bb-pEmpty) or with the BB3576 gene (Bb-BB3576) were measured and expressed relative to Bb-pEmpty vector diameter after 18 h of incubation at 37 °C on SS motility plates supplemented with 32 mM magnesium sulfate. One hundred per cent motility for Bb-pEmpty was 12 mm. The results are based on three replicates. * P<0.01 vs Bb-pEmpty. (c) Biofilm development by Bb-BB3576 compared with Bb-pEmpty was assessed by microtitre plate assays as described in Methods. Biofilm level was determined in a ΔBvgA background (BbΔBvgA-BB3576 and BbΔBvgA-pEmpty). * P<0.01 vs Bb-pEmpty grown at the same modulator concentration, ** P<0.01 vs BbΔBvgA-pEmpty grown at the same modulator concentration.
As was observed for Bb-DGC, overexpression of BB3576 should inhibit swimming motility in Bvgminus phase. The BB3576 gene was cloned into a broad-host-range plasmid pBB1RMCS-5 under control of the nptII constitutive promoter and transferred to Bb-DGC to obtain Bb-BB3576. These bacteria showed reduced swimming motility in soft agar compared with wild-type B. bronchiseptica transformed with the empty vector (Fig. 7b). This behaviour was also observed when BbΔBvgA transformed with the empty vector was compared with BbΔBvgA–BB3576 (data not shown).
As described above, DGC overexpression correlates with impaired motility and enhanced biofilm formation in B. bronchiseptica. Bb-BB3576 showed impaired swimming in soft agar, hence we decided to evaluate if biofilm formation was different from that of the wild-type strain. As expected for a strain with high c-di-GMP levels, biofilm formation of Bb-BB3576 was enhanced in Bvgi phase (1.0 mM NA) compared with B. bronchiseptica transformed with the empty vector (Fig. 7c). Furthermore, biofilm formation for BbΔBvgA–BB3576 in Bvgi phase was enhanced compared with BbΔBvgA transformed with the empty vector (Fig. 7c).
BB3576 complements a diguanylate cyclase mutant phenotype
The BB3576 protein expressed in B. bronchiseptica resulted in phenotypes consistent with this protein being an active DGC. To confirm that BB3576 has DGC activity, we transformed the P. fluorescens Δ4DGC (PsΔ4) strain with the pBB3576 plasmid. Strain PsΔ4 has the genes coding for four DGC proteins deleted from its genome and is not able to produce biofilm under static growth conditions (Newell et al., 2011). A previous study showed that complementation with one DGC protein was sufficient to restore biofilm formation to this strain (Newell et al., 2011). Biofilm formation for the P. fluorescens strains was evaluated in a 96-well plate grown statically for 6 h at 30 °C in K10T-1 medium (Fig. 8). When PsΔ4 was complemented with a DGC-encoding-gene gcbB, biofilm formation was restored as previously described (Newell et al., 2011). When a plasmid carrying BB3576 was introduced, biofilm formation was also observed, suggesting a DGC activity for this protein in concordance with the results described above (Fig. 8).
Fig. 8.
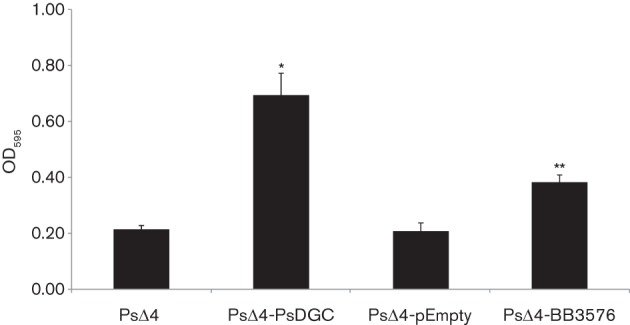
BB3576 complements biofilm phenotype in a P. fluorescens diguanylate cyclase mutant. Biofilm development by the P. fluorescensΔ4DGC strain and P. fluorescensΔ4DGC complemented with BB3576-expressing vector (PsΔ4-BB3576) was determined by CV staining after 6 h growth in static conditions in K10T-1 medium. P. fluorescens with expression vector without insertion (PsΔ4-pEmpty) was used as negative control and P. fluorescens complemented with vector expressing a P. fluorescens diguanylate cyclase gcbB (PsΔ4-PsDGC) as a positive control. gcbB expression was induced by presence of arabinose 0.2 % (p/v). * P<0.001 vs PsΔ4. ** P<0.001 vs PsΔ4-pEmpty.
Discussion
The signalling molecule c-di-GMP has been reported to regulate phenotypes like biofilm formation, motility and virulence. It is generated from two GTP molecules by GGDEF-domain-containing DGC enzymes, and degraded by PDE enzymes containing either EAL or HD-GYP protein domains. Despite the frequent occurrence of these protein domains encoded in bacteria genomes, information on mechanisms and physiological roles regarding their function and regulation is sparse. Many of these proteins have also other domains, suggesting that c-di-GMP activity is regulated by these additional domain functions. So we can imagine that cell c-di-GMP turnover is regulated through the consensus of various external and internal factors. The balance of these signals ultimately regulates biofilm formation, motility and virulence.
In the present work we showed that c-di-GMP signalling is involved in B. bronchiseptica biology. We overexpressed proteins with demonstrated in vitro and in vivo activity in order to establish if c-di-GMP regulation was present in this microbe. We overexpressed PA1120 or PA3947 with known DGC and PDE activities, respectively, in wild-type B. bronchiseptica. As shown in Fig. 1, PA1120 with a GGDEF functional domain overexpression leads to an enhanced biofilm phenotype compared with the wild-type strain under all conditions tested. Biofilm formation was quantified by the CV method and confirmed by SEM. These findings suggest that an increase in the levels of this DGC and in intracellular c-di-GMP levels causes an enhanced production of factors that promote bacterial binding to abiotic surfaces and/or inter-bacterial adherence.
As mentioned above, biofilm formation is regulated by BvgAS in B. bronchiseptica. Nicotinic acid or magnesium sulfate can modulate BvgAS activity in vitro through the three phases, virulent (Bvgplus), intermediate (Bvgi) and avirulent (Bvgminus). Interestingly, although for all modulator conditions Bb-DGC showed more biofilm formation than the parental strain, the difference is bigger in Bvgminus phase (4.0 mM NA). Irie et al. (2004) suggested that while AC activity inhibits biofilm formation, FHA is necessary for full biofilm development. Both proteins are present in Bvgplus phase but only FHA is present in Bvgi phase and both are absent in Bvgminus phase (Mattoo & Cherry, 2005). This is in agreement with results showing that Bvgi phase is the condition where major biofilm formation is observed in B. bronchiseptica. Nevertheless, it is noteworthy that with overexpression of a non-physiological DGC, and in the absence of FHA during growth in the Bvgminus phase, a biofilm is still established by the Bb-DGC strain. The group of factors involved in biofilm regulation and presumably activated by c-di-GMP might be sufficient for establishment of this phenotype regardless of virulence factors like FHA.
In concordance with this finding, when PA1120 was overexpressed in a ΔBvgA background, biofilm formation was observed. However, the biofilm magnitude was significantly weaker than in wild-type background and SEM analysis showed only biofilm architecture in 1.0 mM NA. These results are surprising, since neither BbΔBvgA nor BbΔBvgA-DGC is expected to respond to different concentrations of known BvgAS modulators, thus the interplay between NA and the c-di-GMP network may be present in B. bronchiseptica through mechanisms not yet explored. We can speculate that BvgAS regulates the c-di-GMP network regulating protein expression, particularly those proteins directly involved in c-di-GMP signalling. Interestingly, the BvgR regulatory protein, which represses the expression of virulence genes in Bvgplus phase in Bordetella, contains an EAL domain (Merkel et al., 1998). The expression of this protein is stimulated by the BvgAS system, strongly suggesting a relationship between the BvgAS system and c-di-GMP that requires further investigation.
In other bacteria where c-di-GMP function has been determined, low c-di-GMP levels are frequently associated in planktonic phenotypes with flagellar expression. As shown above, DGC overexpression in B. bronchiseptica enhanced biofilm formation, probably as a consequence of high c-di-GMP levels, which may also repress motility in Bb-DGC when a flagellar system is present. It is known that B. bronchiseptica shows motility in Bvgminus-modulated conditions like high-sulfate concentrations. Motility assays in soft agar plates showed that Bb-DGC exhibited significantly diminished motility. This result is in concordance with behaviour of other bacteria and with a c-di-GMP network that regulates the transition between sessile and motile lifestyles of bacteria. Moreover, we described for the first time, to our knowledge, in B. bronchiseptica a protein with a GGDEF domain that probably had diguanylate cyclase activity in vivo. Real-time PCR results clearly demonstrated that BB3576 expression was higher in Bvgminus phase compared with Bvgplus conditions. However, NA regulation appears to be dominant over BB3576 activity for biofilm formation, even in a ΔBvgA background, since differences in biofilm quantification of strains overexpressing BB3576 were only significant at NA concentrations inducing the Bvgi phase. One possible explanation is that conformational changes in the expressed BB3576 protein may be required for optimal DGC function. BB3576 has two predicted domains, a DGC domain and an extracellular sensory Cache domain (Ca+2 channels, chemotaxis receptors). In general, Cache domains sense stimuli present in the periplasm and transmit signals to an output domain such as GGDEF, resulting in a specific adaptive response (Anantharaman & Aravind 2000; Galperin 2006). We speculate that BB3576 regulates biofilm and motility when a ligand binds to the Cache domain. The nature of this ligand remains unknown at the moment. Diguanylate cyclase activity was demonstrated in vivo for BB3576 as shown in Fig. 8. Overexpression of BB3576 in a P. fluorescens strain with a low-DGC-activity background restored biofilm formation. The partial complementation achieved by BB3576 may be explained by the absence from the growth media of the unknown ligand.
Recently, Amarasinghe and co-workers described YeaJ in Salmonella enterica serovar Typhimurium as an active DGC membrane protein involved in motility regulation and exopolysaccharide production (Amarasinghe et al., 2013). This protein, like BB3576, has a GGDEF domain with demonstrated DGC activity and a putative Cache domain. They proposed that the association of a protective monoclonal IgA antibody with the O-antigen induces outer membrane stress triggering a c-di-GMP signalling pathway that effectively promotes a less motile and non-invasive biofilm state, thereby rendering the bacterium unable to invade intestinal epithelial cells. The particular expression profile of BB3576 in Bvgplus, Bvgi and Bvgminus phases is in concordance with putative motility regulation of the bacteria during Bvgminus phase. Flagellar synthesis and function is regulated in bacteria at different levels by c-di-GMP (Wolfe & Visick, 2008). This newly described protein BB3576 might fine tune flagellar motor function using c-di-GMP as a second messenger in response to chemosensory activity of the Cache domain, as reported for E. coli (Pesavento et al., 2008).
Although further research is needed to elucidate c-di-GMP regulation and signalling in B. bronchiseptica, we demonstrated here for the first time, to our knowledge, its role regarding the control of B. bronchiseptica biofilm formation and motility. More over we would now argue that c-di-GMP is involved in motility regulation in response to as yet unknown ligands that may be sensed by BB3576.
Acknowledgements
We thank to Stephen Lory for the plasmids containing PA1120 and PA3947 genes. We acknowledge the Servicio de Microscopía Electrónica de Barrido [Centro de Investigación y Desarrollo en Procesos Catalíticos (CINDECA)–Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET)–Universidad Nacional de La Plata (UNLP)] and Laboratorio de Investigaciones de Metalurgia Física (LIMF) service (Facultad de Ingeniería–UNLP) for SEM analysis. This work was supported by the Agencia Nacional de Promoción Científica y Tecnológica ANCPyT and CONICET [PIP N°0920 and Cooperación Internacional CONICET–National Science foundation (NSF)] grants to D. F. H., F. S. and J. F, National Institutes for Health (NIH) grant R01A1003256 to G. A. O. and a Rosalind Borrison Fellowship to D. G. H. D .F. H. is a member of the Scientific Career of the Comisión de Investigaciones Científicas de la Provincia de Buenos Aires (CICBA). J. F. and F. S. are members of the Scientific Career of CONICET.
Abbreviations:
- AC
adenylate cyclase
- c-di-GMP
bis-(3′–5′)-cyclic-dimeric guanosine monophosphate
- Ct
threshold cycle
- CV
crystal violet
- DGC
diguanylate cyclase
- FHA
filamentous haemagglutinin
- NA
nicotinic acid
- PDE
phosphodiesterase
- PRN
pertactin
- SEM
scanning electron microscopy
- TTSS
type three secretory system
Footnotes
Two supplementary tables, listing plasmids and strains used and primers used in PCRs, are available with the online version of this paper.
References
- Akerley B. J., Miller J. F. (1993). Flagellin gene transcription in Bordetella bronchiseptica is regulated by the BvgAS virulence control system. J Bacteriol 175, 3468–3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akerley B. J., Monack D. M., Falkow S., Miller J. F. (1992). The bvgAS locus negatively controls motility and synthesis of flagella in Bordetella bronchiseptica. J Bacteriol 174, 980–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amarasinghe J. J., D’Hondt R. E., Waters C. M., Mantis N. J. (2013). Exposure of Salmonella enterica Serovar Typhimurium to a protective monoclonal IgA triggers exopolysaccharide production via a diguanylate cyclase-dependent pathway. Infect Immun 81, 653–664. 10.1128/IAI.00813-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amikam D., Galperin M. Y. (2006). PilZ domain is part of the bacterial c-di-GMP binding protein. Bioinformatics 22, 3–6. 10.1093/bioinformatics/bti739 [DOI] [PubMed] [Google Scholar]
- Anantharaman V., Aravind L. (2000). Cache – a signaling domain common to animal Ca2+-channel subunits and a class of prokaryotic chemotaxis receptors. Trends Biochem Sci 25, 535–537. 10.1016/S0968-0004(00)01672-8 [DOI] [PubMed] [Google Scholar]
- Conover M. S., Mishra M., Deora R. (2011). Extracellular DNA is essential for maintaining Bordetella biofilm integrity on abiotic surfaces and in the upper respiratory tract of mice. PLoS ONE 6, e16861. 10.1371/journal.pone.0016861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotter P. A., Miller J. F. (1997). A mutation in the Bordetella bronchiseptica bvgS gene results in reduced virulence and increased resistance to starvation, and identifies a new class of Bvg-regulated antigens. Mol Microbiol 24, 671–685. 10.1046/j.1365-2958.1997.3821741.x [DOI] [PubMed] [Google Scholar]
- Dombrecht B., Vanderleyden J., Michiels J. (2001). Stable RK2-derived cloning vectors for the analysis of gene expression and gene function in Gram-negative bacteria. Mol Plant Microbe Interact 14, 426–430. 10.1094/MPMI.2001.14.3.426 [DOI] [PubMed] [Google Scholar]
- Fernández J., Sisti F., Bottero D., Gaillard M. E., Hozbor D. (2005). Constitutive expression of bvgR-repressed factors is not detrimental to the Bordetella bronchiseptica–host interaction. Res Microbiol 156, 843–850. 10.1016/j.resmic.2005.04.003 [DOI] [PubMed] [Google Scholar]
- Galperin M. Y. (2004). Bacterial signal transduction network in a genomic perspective. Environ Microbiol 6, 552–567. 10.1111/j.1462-2920.2004.00633.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galperin M. Y. (2005). A census of membrane-bound and intracellular signal transduction proteins in bacteria: bacterial IQ, extroverts and introverts. BMC Microbiol 5, 35. 10.1186/1471-2180-5-35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galperin M. Y. (2006). Structural classification of bacterial response regulators: diversity of output domains and domain combinations. J Bacteriol 188, 4169–4182. 10.1128/JB.01887-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galperin M. Y., Nikolskaya A. N., Koonin E. V. (2001). Novel domains of the prokaryotic two-component signal transduction systems. FEMS Microbiol Lett 203, 11–21. 10.1111/j.1574-6968.2001.tb10814.x [DOI] [PubMed] [Google Scholar]
- Goodnow R. A. (1980). Biology of Bordetella bronchiseptica. Microbiol Rev 44, 722–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvill E. T., Cotter P. A., Yuk M. H., Miller J. F. (1999). Probing the function of Bordetella bronchiseptica adenylate cyclase toxin by manipulating host immunity. Infect Immun 67, 1493–1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvill E. T., Preston A., Cotter P. A., Allen A. G., Maskell D. J., Miller J. F. (2000). Multiple roles for Bordetella lipopolysaccharide molecules during respiratory tract infection. Infect Immun 68, 6720–6728. 10.1128/IAI.68.12.6720-6728.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengge R. (2009). Principles of c-di-GMP signalling in bacteria. Nat Rev Microbiol 7, 263–273. 10.1038/nrmicro2109 [DOI] [PubMed] [Google Scholar]
- Inatsuka C. S., Xu Q., Vujkovic-Cvijin I., Wong S., Stibitz S., Miller J. F., Cotter P. A. (2010). Pertactin is required for Bordetella species to resist neutrophil-mediated clearance. Infect Immun 78, 2901–2909. 10.1128/IAI.00188-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irie Y., Mattoo S., Yuk M. H. (2004). The Bvg virulence control system regulates biofilm formation in Bordetella bronchiseptica. J Bacteriol 186, 5692–5698. 10.1128/JB.186.17.5692-5698.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenal U., Malone J. (2006). Mechanisms of cyclic-di-GMP signaling in bacteria. Annu Rev Genet 40, 385–407. 10.1146/annurev.genet.40.110405.090423 [DOI] [PubMed] [Google Scholar]
- Kolter R., Greenberg E. P. (2006). Microbial sciences: the superficial life of microbes. Nature 441, 300–302. 10.1038/441300a [DOI] [PubMed] [Google Scholar]
- Kovach M. E., Elzer P. H., Hill D. S., Robertson G. T., Farris M. A., Roop R. M., II, Peterson K. M. (1995). Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166, 175–176. 10.1016/0378-1119(95)00584-1 [DOI] [PubMed] [Google Scholar]
- Kulesekara H., Lee V., Brencic A., Liberati N., Urbach J., Miyata S., Lee D. G., Neely A. N., Hyodo M., et al. (2006). Analysis of Pseudomonas aeruginosa diguanylate cyclases and phosphodiesterases reveals a role for bis-(3′–5′)-cyclic-GMP in virulence. Proc Natl Acad Sci U S A 103, 2839–2844. 10.1073/pnas.0511090103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai T. H., Kumagai Y., Hyodo M., Hayakawa Y., Rikihisa Y. (2009). The Anaplasma phagocytophilum PleC histidine kinase and PleD diguanylate cyclase two-component system and role of cyclic di-GMP in host cell infection. J Bacteriol 191, 693–700. 10.1128/JB.01218-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattoo S., Cherry J. D. (2005). Molecular pathogenesis, epidemiology, and clinical manifestations of respiratory infections due to Bordetella pertussis and other Bordetella subspecies. Clin Microbiol Rev 18, 326–382. 10.1128/CMR.18.2.326-382.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillan D. J., Shojaei M., Chhatwal G. S., Guzmán C. A., Walker M. J. (1996). Molecular analysis of the bvg-repressed urease of Bordetella bronchiseptica. Microb Pathog 21, 379–394. 10.1006/mpat.1996.0069 [DOI] [PubMed] [Google Scholar]
- Merkel T. J., Barros C., Stibitz S. (1998). Characterization of the bvgR locus of Bordetella pertussis. J Bacteriol 180, 1682–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen H., Ball G., Giraud C., Filloux A. (2009). Expression of Pseudomonas aeruginosa CupD fimbrial genes is antagonistically controlled by RcsB and the EAL-containing PvrR response regulators. PLoS ONE 4, e6018. 10.1371/journal.pone.0006018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra M., Parise G., Jackson K. D., Wozniak D. J., Deora R. (2005). The BvgAS signal transduction system regulates biofilm development in Bordetella. J Bacteriol 187, 1474–1484. 10.1128/JB.187.4.1474-1484.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newell P. D., Yoshioka S., Hvorecny K. L., Monds R. D., O’Toole G. A. (2011). Systematic analysis of diguanylate cyclases that promote biofilm formation by Pseudomonas fluorescens Pf0-1. J Bacteriol 193, 4685–4698. 10.1128/JB.05483-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesavento C., Becker G., Sommerfeldt N., Possling A., Tschowri N., Mehlis A., Hengge R. (2008). Inverse regulatory coordination of motility and curli-mediated adhesion in Escherichia coli. Genes Dev 22, 2434–2446. 10.1101/gad.475808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Römling U., Amikam D. (2006). Cyclic di-GMP as a second messenger. Curr Opin Microbiol 9, 218–228. 10.1016/j.mib.2006.02.010 [DOI] [PubMed] [Google Scholar]
- Sanchez-Torres V., Hu H., Wood T. K. (2011). GGDEF proteins YeaI, YedQ, and YfiN reduce early biofilm formation and swimming motility in Escherichia coli. Appl Microbiol Biotechnol 90, 651–658. 10.1007/s00253-010-3074-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sisti F., Fernández J., Rodríguez M. E., Lagares A., Guiso N., Hozbor D. F. (2002). In vitro and in vivo characterization of a Bordetella bronchiseptica mutant strain with a deep rough lipopolysaccharide structure. Infect Immun 70, 1791–1798. 10.1128/IAI.70.4.1791-1798.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner J. A., Reissinger A., Shen H., Yuk M. H. (2004). Bordetella type III secretion and adenylate cyclase toxin synergize to drive dendritic cells into a semimature state. J Immunol 173, 1934–1940. [DOI] [PubMed] [Google Scholar]
- Sloan G. P., Love C. F., Sukumar N., Mishra M., Deora R. (2007). The Bordetella Bps polysaccharide is critical for biofilm development in the mouse respiratory tract. J Bacteriol 189, 8270–8276. 10.1128/JB.00785-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stainer D. W., Scholte M. J. (1970). A simple chemically defined medium for the production of phase I Bordetella pertussis. J Gen Microbiol 63, 211–220. [DOI] [PubMed] [Google Scholar]
- Stockbauer K. E., Fuchslocher B., Miller J. F., Cotter P. A. (2001). Identification and characterization of BipA, a Bordetella Bvg-intermediate phase protein. Mol Microbiol 39, 65–78. 10.1046/j.1365-2958.2001.02191.x [DOI] [PubMed] [Google Scholar]
- Wan X., Tuckerman J. R., Saito J. A., Freitas T. A., Newhouse J. S., Denery J. R., Galperin M. Y., Gonzalez G., Gilles-Gonzalez M. A., Alam M. (2009). Globins synthesize the second messenger bis-(3′–5′)-cyclic diguanosine monophosphate in bacteria. J Mol Biol 388, 262–270. 10.1016/j.jmb.2009.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams C. L., Cotter P. A. (2007). Autoregulation is essential for precise temporal and steady-state regulation by the Bordetella BvgAS phosphorelay. J Bacteriol 189, 1974–1982. 10.1128/JB.01684-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe A. J., Visick K. L. (2008). Get the message out: cyclic-di-GMP regulates multiple levels of flagellum-based motility. J Bacteriol 190, 463–475. 10.1128/JB.01418-07 [DOI] [PMC free article] [PubMed] [Google Scholar]