Abstract
PubServer, available at http://pubserver.burnham.org/, is a tool to automatically collect, filter and analyze publications associated with groups of homologous proteins. Protein entries in databases such as Entrez Protein database at NCBI contain information about publications associated with a given protein. The scope of these publications varies a lot: they include studies focused on biochemical functions of individual proteins, but also reports from genome sequencing projects that introduce tens of thousands of proteins. Collecting and analyzing publications related to sets of homologous proteins help in functional annotation of novel protein families and in improving annotations of well-studied protein families or individual genes. However, performing such collection and analysis manually is a tedious and time-consuming process. PubServer automatically collects identifiers of homologous proteins using PSI-Blast, retrieves literature references from corresponding database entries and filters out publications unlikely to contain useful information about individual proteins. It also prepares simple vocabulary statistics from titles, abstracts and MeSH terms to identify the most frequently occurring keywords, which may help to quickly identify common themes in these publications. The filtering criteria applied to collected publications are user-adjustable. The results of the server are presented as an interactive page that allows re-filtering and different presentations of the output.
INTRODUCTION
The rapid expansion of molecular biology databases storing sequences, structures, results of high-throughput experiments and literature creates a growing need for establishing links between different types of information (for broad discussion and insights into this subject, please see recent studies (1,2)). In particular, protein and gene entries deposited in public databases such as Uniprot (3) or GenBank (4) contain curated or depositor-supplied, respectively, links to related publications. Several specialized resources, such as GeneCards (5), OMIM (6), MGD (7), EcoCyc (8) and many others also collect publications about individual genes or proteins from specific organisms. Protein family databases such as Pfam (9) annotate proteins as members of families and provide short descriptions for most of them. However, almost one-third of all Pfam families lack such descriptions and are annotated as ‘domains of unknown function’ (DUFs) (10). Researchers interested in finding information about a specific protein can use all these resources to collect peer-reviewed manuscripts providing information about their protein or gene of interest. However, the task of manual collection and review of literature about proteins is a time-consuming process, involving sequence similarity searches, opening and reading tens or hundreds of database entries, collecting literature references listed in these entries and eliminating publications that only describe sequencing projects and other large-scale studies. When we expand this task to an entire protein family, it becomes almost prohibitively time-consuming. Moreover, collecting literature from curated database entries provides excellent results for well-studied proteins and protein families, but is usually less effective for uncharacterized ones.
These situations led to the development of methods that use a protein sequence as a starting point to query literature databases (11–15) (see Table 1). For instance, METIS (14) and GeneReporter (11) retrieve publications listed on annotated UNIPROT or SWISSPROT pages. The quickLit (13) and METIS servers rely on a fast, but less-sensitive sequence–sequence comparison methods (BlastP and BlastX (16), and BlastP, respectively). The GeneReporter server uses a more-sensitive PSI-Blast (17) algorithm. Our tests of DUF families suggested that, while these services provide a lot of useful information for proteins from some protein families, in some cases relevant publications can be found only by exhaustive PSI-Blast searches of the complete NR database followed by the analysis of the Entrez entries for collected proteins.
Table 1.
Comparison of services for sequence-based literature retrieval
| Server's name and URL | Sequence database(s) | Search method(s) | Reference |
|---|---|---|---|
| GeneReporter | UNIPROT | BlastP, WU-Blast, PSI-Blast | MineBlast (12) |
| (replaced MineBlast) | GeneReporter (11) | ||
| http://www.genereporter.tu-bs.de/ | |||
| METIS | SWISSPROT | BlastP | (14) |
| http://www.bioinf.manchester.ac.uk/cgi-bin/dbbrowser/precis/metis_precis.cgi | |||
| quickLit | Proteomes of seven model organisms | BlastP, BlastX | (13) |
| http://genomics.nimr.mrc.ac.uk/apps/quickLit/ | |||
| PubServer | NR, (including SWISSPROT) | PSI-Blast | |
| http://pubserver.burnham.org/ |
In order to enable such broad searches and to complement existing methods, we developed PubServer, which automatically gathers and filters literature listed in protein Entrez pages for a list of homologous proteins identified by a PSI-Blast search. Performing such tasks automatically relieves researchers from the most time-consuming elements of a massive literature search. For uncharacterized proteins, PubServer may help to find a publication that provides some hypothesis about the proteins' functions, and, for well-studied ones, it may often collect additional literature that could not be found easily by text-based searches. For well-characterized proteins, sequence-based literature retrieval complements traditional keyword-based searches by addressing the problem of multiple synonyms and nomenclature differences between organisms. However, sequence-based literature search methods such as PubServer are not function prediction algorithms, replacements for traditional text-based literature searches or substitutes for specialized gene annotation resources. Because of its large database and sensitive homology recognition method, PubServer may be especially useful in finding clues about functions of uncharacterized protein families for which a remote similarity to an annotated protein sometimes provides the only hint about their function.
OVERVIEW OF THE SERVER
The input to PubServer is a protein sequence entered as text in the entry form on the first page of the server. In the next step, the server performs a PSI-Blast (17) search against the NR database, using parameters that can be modified by the user. The resulting list of homologs (internally represented by GI numbers) is processed to identify a list of associated publications (represented by PubMed (18,19) ids) using a precalculated protein-to-publication mapping table. Then, publications referring to large numbers of proteins are filtered out from the list according to the user-defined cutoff (i.e. the maximum number of proteins to which a publication may refer to). In the next step, PubServer collects words used in titles, abstracts and MeSH (20) terms of the gathered publications and prepares a summary list of used vocabulary. The resulting vocabulary list may provide clues about common themes found in collected publications and can also be used as a starting point for additional filtering. However, it does not reflect the precise distribution of scientific terms found in the publications (also see point 5 below). It also has to be noted that PubServer can only collect publications listed as references on Entrez pages of sequences deposited in GeneBank or transferred from SWISSPROT (for instance, PubServer would not find the review about a protein family, unless it is listed in the Entrez database entry of at least one protein from this family).
PUBSERVER WORKFLOW
Input
The input for PubServer is an amino-acid sequence of a protein of interest in fasta format. The server accepts sequences containing up to 5000 residues; however, it is recommended to split sequences longer than about 500 residues into domains. The input form and parameters of the server are shown in Figure 1A.
Figure 1.
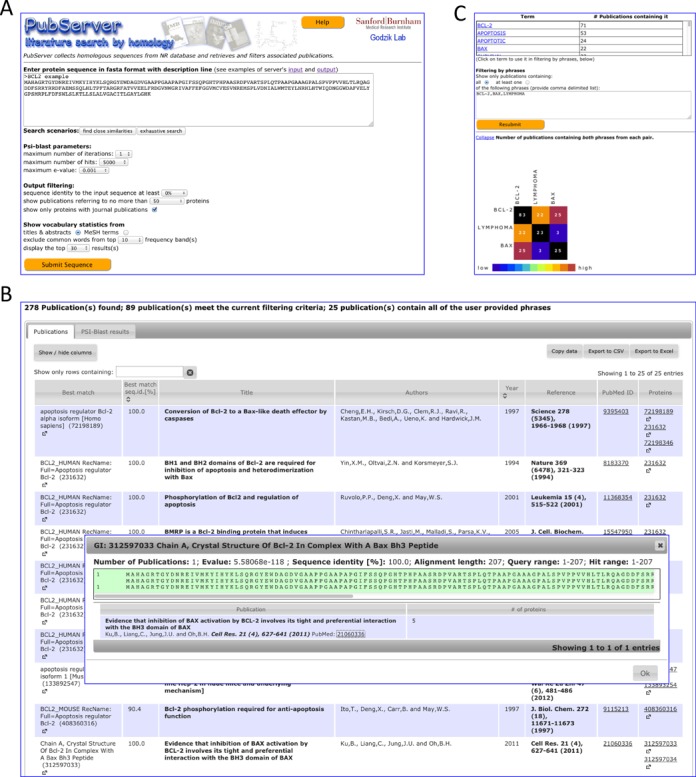
(A) PubServer input form. (B) Output publication list. Detailed information about query–hit similarity and alignment field is displayed in a pop-up window. (C) Vocabulary statistics and filtering interface elements. Phrases to be used in filtering can be copied with one click from the vocabulary list or entered and edited manually. PubServer can show publications containing ‘at least one’ or ‘all’ of the entered terms. A simple matrix illustrating co-occurrence of entered phrases in publications is also displayed.
Calculations
PubServer performs all calculations on a local Linux cluster. A typical job takes between 3 and 30 min—depending on the sequence length and the number of homologs found—and consists of the following steps:
Collect sequences of proteins homologous to the input protein from the NR database using PSI-Blast. PSI-Blast parameters such as the number of iterations, the e-value cutoff and the maximum number of collected sequences are user-adjustable. PubServer also allows filtering collected sequences according to their sequence identity to the query. Two sets of preset parameter values corresponding to the two typical search scenarios are available by clicking ‘find close similarities’ or ‘exhaustive search’ buttons (also see section ‘Suggestions for PubServer use’ below).
Collect publications from the database entries of proteins found using the PSI-Blast search. All entries corresponding to different depositions of the same protein are included; thus, a full REDUNDANT Entrez database is analyzed. For better performance, the search is done using a mapping table linking all protein entries (represented as GI numbers) to publications (represented as PubMed IDs). This table, prepared and regularly updated based on all protein Entrez pages downloaded from NCBI, is stored on the server.
Filter collected publications according to the number of proteins to which they refer in order to eliminate publications about sequencing projects and other large-scale studies (the cutoff number of proteins a publication can refer to is adjustable).
Parse titles, abstracts and MeSH terms from the collected publications and prepare the list of vocabulary statistics.
Filter out common English words from the collected vocabulary list with an adjustable filter based on the 2+2gfreq (21) word list, which contains English vocabulary divided into 27 frequency bands. The first band contains the most commonly found words (i.e. ‘the,’ ‘and,’ ‘to’) that obviously do not carry any specific information about protein function and should not be included in the vocabulary list. On the other hand, the bottom frequency bands contain specialized and potentially informative words. In our experience, excluding the top 10 bands from the vocabulary analysis is a good starting point, and it is set as an initial value on the server. However, it has to be noted that, besides removing the most common words based on a simple frequency criterion, the vocabulary list is not processed by merging synonyms, stemming or matching it to any predefined dictionary. Thus, the vocabulary list is only an approximation of the distribution of biological terms found in the titles, abstracts or MeSH terms of collected publications.
Optionally filter the publication list according to the presence of words (or phrases) found in the titles and abstracts. The phrases to be used in filtering the publication list can be copied from the vocabulary list (simply by clicking on them) or can be entered manually. If filtering by phrases is performed, then simple co-occurrence statistics of vocabulary elements used in filtering are also calculated and presented as a matrix. The output may also be narrowed down to publications containing at least one or all of the user-provided phrases. After resubmitting the form, PubServer would filter the collected publications and show an expandable field with a simple co-occurrence matrix illustrating distribution of these terms in the collected publications (see Figure 1C).
Present the PSI-Blast results and Publications lists as sortable tables with an additional fast text filtering option.
Re-filtering of the output
The output interface allows changes of parameters used in filters described in steps 3–7 above without rerunning a PSI-Blast search, which is only necessary if PSI-Blast parameters are to be changed.
Output
The output of PubServer is composed of two tables displayed on two tabs. The first tab, called ‘Publications,’ contains a list of collected publications initially ranked by the maximum sequence identity between the query sequence and the closest sequence found by PSI-Blast (and linked to the publications). The table columns include: Best match (protein description), Best match e-value, Best match seq. id. [%], Title, Authors, Year, Reference, PubMed ID (and a link to the publication's abstract), MeSH terms, Number of proteins (to which the publication refers), Number of proteins in PSI-Blast output and Proteins (list of GI numbers and links to Entrez pages of these proteins) (see Figure 1B). The table allows hiding and showing of selected columns, sorting and fast filtering by text (i.e. limiting the display to rows containing specific text or number). Clicking on the protein's GI number displays a pop-up window with the protein's description (if available) and parameters of the PSI-Blast alignment between the query sequence and the protein. The pop-up window also contains the alignment and a full list of publications listed on the protein's Entrez page.
The second tab, called ‘PSI-Blast results,’ contains the list of proteins found by PSI-Blast presented as a table. The columns include GI, Description (if available), E-value, Sequence identity [%], Alignment length, Query start, Query end, Hit start, Hit end, PSI-Blast iteration, Number of publications (listed on the protein's Entrez page) and PubMed IDs (links to publication pages in PubMed). Clicking on a protein's GI number opens a protein pop-up window described in the previous paragraph.
SUGGESTIONS FOR PUBSERVER USE
Input parameters for different applications of PubServer
The PubServer input form contains adjustable parameters that make it possible to customize the search for a specific purpose.
At one extreme of the protein similarity range, finding literature related to well-studied orthologs of human proteins in other vertebrates would usually require a single iteration of PSI-Blast with an e-value cutoff of 10−10 or 10−20. Optimal cutoff values for sequence identity would vary between different proteins, but the suggested starting value may be 0%. Since the sequence identity threshold can be modified and quickly reapplied to already-calculated PSI-Blast results, it might be better to test a few sequence identity cutoff values and examine the resulting publication list. The list may be additionally filtered using phrases found in the titles, abstracts and MeSH terms. The suggested initial parameters for this search scenario can be quickly set by clicking the ‘find close similarities’ button.
At the opposite extreme of the protein similarity range, the question of finding ‘any’ information about ‘any’ homolog of an uncharacterized protein or a member of a protein family of unknown function sometimes requires exhaustive PSI-Blast searches with five or more iterations and an e-value cutoff of 10−3. The initial value of the sequence identity threshold should be set at 0%, and, only if a substantial number of informative publications are found, it may be increased in order to select literature about the closest homologs of a query. The suggested initial parameters for this search scenario can be quickly set by clicking the ‘exhaustive search’ button.
It should be noted that, while PSI-Blast searches rarely produce false positives when used with conservative e-value cutoffs and we can expect that all identified proteins are indeed homologous to the query, this set would often extend beyond the scope of a protein family as formally defined in resources such as Pfam. The functional information derived from homologous proteins that formally belong to another (related) protein family may still be informative; however, in such cases, one cannot say that all collected publications describe ‘members’ of the original protein family of interest.
Annotating protein families and protein domains
The term ‘protein family’ used several times in this text may refer to a group of related ‘full-length’ proteins or to a protein domain (a group of related ‘regions’ of proteins that, in individual proteins, may be accompanied by other domains). Obviously, PubServer searches aimed at proposing a hypothesis about the function of a protein domain should be seeded with a subsequence representing an instance of this domain. However, one should still bear in mind that collected publications are usually associated with full-length proteins, rather than with individual domains. For example, if the output list contains publications describing DNA-binding proteins, it does not imply that our domain of interest binds DNA but only that it is found in some number of proteins that bind DNA. Therefore, proposing a hypothesis about a domain's function often requires checking alignments, domain architectures of collected proteins and collected publications. Obviously, if proteins associated with informative publications do not contain other domains, then the function of these proteins is a good hypothesis for the domain's function.
EXAMPLES OF PubServer APPLICATIONS
The following examples illustrate how PubServer can be used to quickly collect publications helpful in proposing hypotheses about functions of uncharacterized protein families. In two of these examples besides PubServer, we used a separate remote homology prediction algorithm, FFAS03 (22) since using more than one independent source of functional information leads to a more solid hypothesis.
Establishing function and collecting literature for proteins from the DUF659 and DUF4371 families
Protein families DUF659 and DUF4371 are linked by a marginal homology detected with the profile–profile alignment algorithm FFAS03. Both families also show a weak similarity to the catalytic (DNA-binding) domain from hermes DNA transposase (PDB ID: 2bw3), but they currently do not have a functional annotation in the Pfam database. PubServer searches seeded with sequences of members of any of these families reveal several publications describing individual proteins as transposases. Based on this information, one can hypothesize that these two protein families most likely consist of DNA-binding domains of transposases.
Identification of a functionally annotated member of a protein family of unknown function, DUF2522
This protein family contains about 150 proteins from the Bacilli class of bacteria. It is not currently annotated in the Pfam database, and remote homology detection methods do not reveal any significant similarity to annotated protein families or structures. However, a PubServer search seeded with a representative of this family immediately identified a publication focused specifically on one family member that was described as a sporulation protein inhibiting DNA replication (23). It has to be noted that a direct PSI-Blast search in the NR database identifies at least 10 proteins with annotations suggesting a link to sporulation (among over 100 ‘hypothetical’ proteins). However, finding out whether any of these proteins was actually experimentally characterized and whether it was published requires manual checking. PubServer allowed immediate identification of the experimentally characterized proteins and the associated publications.
Collecting literature about members of a family of unknown function, DUF3380
This protein family contains over 200 proteins from bacteria and phages and does not have a functional annotation in the Pfam database. However, the Pfam page for this family shows that this domain is often found in combination with a peptidoglycan-binding domain (although it is also found in one-domain proteins). Remote homology detection methods such as FFAS03 show that it is marginally similar to lysozyme, chitinase and other cell wall hydrolases. Many proteins from this family are actually annotated as peptidoglycan-binding proteins and bacteriophage-acquired proteins; however, since in different projects, annotations are often transferred between homologs, it is not clear if any of these proteins are directly experimentally characterized. A PubServer search started with three PSI-Blast iterations collected 94 publications, and, after filtering out publications referring to more than 50 proteins, it produced a list of seven references. As immediately indicated by vocabulary statistics collected by PubServer and then confirmed by reviewing these publications, almost all of them describe members of DUF3380 as cell-wall-peptidoglycan lytic enzymes. Because remote homology to proteins with similar functions and direct evidence collected by PubServer point in the same direction, we can, with a relatively high level of confidence, annotate DUF3380 as a family of cell-wall lytic enzymes. It is also noteworthy that a direct PubMed search with the keyword ‘DUF3380’ did not retrieve any of these publications.
Collecting and filtering literature associated with deposited sequences of human antibodies
For very large and diverse proteins families, PubServer provides an additional way of collecting and filtering literature. For instance, one can collect literature associated with protein Entrez entries of immunoglobulins and then retrieve only publications linked to depositions of sequences of human antibodies. In that case, an initial PSI-Blast search collects thousands of immunoglobulin sequences associated with hundreds of publications, but filtering by keywords makes it possible to narrow them down to human antibodies (by retrieving publications containing both keywords ‘human’ and ‘antibody’). If the original PubServer query was seeded with a sequence of a specific human antibody of interest, then the output may be also limited to antibodies sharing a certain level of sequence identity with that protein.
SUMMARY
The volume of biomedical literature stored in public databases is expanding at an exponential pace, creating new challenges for literature mining (19). Similar rapid growth is observed for sequence information and other results of high-throughput experiments, creating a complex challenge of linking information from different specialized databases (1,2). Despite substantial progress in automated text analysis and efforts toward introducing consistent ontologies for biological terminology (24), finding all literature related to a given protein or family of proteins is often a nontrivial task marred by the existence of multiple synonymous gene names and other differences in terminology. Moreover, standard literature searches based on keywords cannot be performed for uncharacterized and, thus, usually unnamed proteins. By searching literature by sequence similarity rather than by keywords, PubServer provides a useful supplement to traditional text-based searches.
Currently, the biggest limitation of PubServer is the fact that it collects only publications listed in Entrez Protein database entries (i.e. publications linked to the deposition of the protein or nucleotide sequence). We are planning to expand PubServer by supplementing currently used protein–publication association lists retrieved from NCBI protein pages with additional lists prepared based on the text mining of available biomedical literature.
FUNDING
National Institute of General Medical Sciences of the National Institutes of Health (NIH) [U54 GM094586]. Funding for open access charge: NIH [U54 GM094586] and funds from Sanford Burnham Medical Research Institute.
Conflict of interest statement. None declared.
REFERENCES
- 1.Kafkas S., Kim J.H., McEntyre J.R. Database citation in full text biomedical articles. PloS One. 2013;8:e63184. doi: 10.1371/journal.pone.0063184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Neveol A., Wilbur W.J., Lu Z. Improving links between literature and biological data with text mining: a case study with GEO, PDB and MEDLINE. Database. 2012;2012:bas026. doi: 10.1093/database/bas026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Activities at the Universal Protein Resource (UniProt) Nucleic Acids Res. 2014;42:D191–D198. doi: 10.1093/nar/gkt1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benson D.A., Cavanaugh M., Clark K., Karsch-Mizrachi I., Lipman D.J., Ostell J., Sayers E.W. GenBank. Nucleic Acids Res. 2013;41:D36–D42. doi: 10.1093/nar/gks1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Safran M., Dalah I., Alexander J., Rosen N., Iny Stein T., Shmoish M., Nativ N., Bahir I., Doniger T., Krug H., et al. GeneCards Version 3: the human gene integrator. Database. 2010;2010:baq020. doi: 10.1093/database/baq020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Amberger J., Bocchini C.A., Scott A.F., Hamosh A. McKusick's Online Mendelian Inheritance in Man (OMIM) Nucleic Acids Res. 2009;37:D793–D796. doi: 10.1093/nar/gkn665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blake J.A., Bult C.J., Eppig J.T., Kadin J.A., Richardson J.E. The Mouse Genome Database: integration of and access to knowledge about the laboratory mouse. Nucleic Acids Res. 2014;42:D810–D817. doi: 10.1093/nar/gkt1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Keseler I.M., Mackie A., Peralta-Gil M., Santos-Zavaleta A., Gama-Castro S., Bonavides-Martinez C., Fulcher C., Huerta A.M., Kothari A., Krummenacker M., et al. EcoCyc: fusing model organism databases with systems biology. Nucleic Acids Res. 2013;41:D605–D612. doi: 10.1093/nar/gks1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Finn R.D., Bateman A., Clements J., Coggill P., Eberhardt R.Y., Eddy S.R., Heger A., Hetherington K., Holm L., Mistry J., et al. Pfam: the protein families database. Nucleic Acids Res. 2014;42:D222–D230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bateman A., Coggill P., Finn R.D. DUFs: families in search of function. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2010;66:1148–1152. doi: 10.1107/S1744309110001685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bartsch A., Bunk B., Haddad I., Klein J., Munch R., Johl T., Karst U., Jansch L., Jahn D., Retter I. GeneReporter–sequence-based document retrieval and annotation. Bioinformatics. 2011;27:1034–1035. doi: 10.1093/bioinformatics/btr047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dieterich G., Karst U., Wehland J., Jansch L. MineBlast: a literature presentation service supporting protein annotation by data mining of BLAST results. Bioinformatics. 2005;21:3450–3451. doi: 10.1093/bioinformatics/bti528. [DOI] [PubMed] [Google Scholar]
- 13.Gilchrist M.J., Christensen M.B., Harland R., Pollet N., Smith J.C., Ueno N., Papalopulu N. Evading the annotation bottleneck: using sequence similarity to search non-sequence gene data. BMC Bioinformatics. 2008;9:442. doi: 10.1186/1471-2105-9-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mitchell A.L., Divoli A., Kim J.H., Hilario M., Selimas I., Attwood T.K. METIS: multiple extraction techniques for informative sentences. Bioinformatics. 2005;21:4196–4197. doi: 10.1093/bioinformatics/bti675. [DOI] [PubMed] [Google Scholar]
- 15.Tu Q., Tang H., Ding D. MedBlast: searching articles related to a biological sequence. Bioinformatics. 2004;20:75–77. doi: 10.1093/bioinformatics/btg375. [DOI] [PubMed] [Google Scholar]
- 16.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 17.Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2014;42:D7–D17. doi: 10.1093/nar/gkt1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu Z. PubMed and beyond: a survey of web tools for searching biomedical literature. Database. 2011;2011:baq036. doi: 10.1093/database/baq036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Joubert M., Fieschi M., Robert J.J., Volot F., Fieschi D. UMLS-based conceptual queries to biomedical information databases: an overview of the project ARIANE. Unified Medical Language System. J. Am. Med. Inform. Assoc. 1998;5:52–61. doi: 10.1136/jamia.1998.0050052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Beale A. Release 5 of the 12dicts word lists. 2007. [Google Scholar]
- 22.Jaroszewski L., Rychlewski L., Li Z., Li W., Godzik A. FFAS03: a server for profile–profile sequence alignments. Nucleic Acids Res. 2005;33:W284–288. doi: 10.1093/nar/gki418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rahn-Lee L., Gorbatyuk B., Skovgaard O., Losick R. The conserved sporulation protein YneE inhibits DNA replication in Bacillus subtilis. J. Bacteriol. 2009;191:3736–3739. doi: 10.1128/JB.00216-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]


