Figure 1.
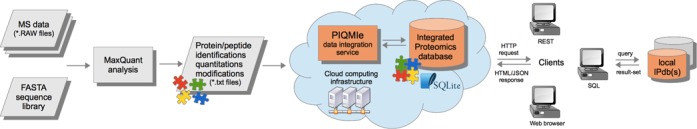
Computational proteomics workflow including the PIQMIe service. Before using the service, the semi-quantitative MS data are analyzed by the MaxQuant software. The resulting files, i.e. the peptide (‘evidence.txt’) and protein lists (‘proteinGroups.txt’) are uploaded together with the used FASTA sequence library to the server through the submission web page. PIQMIe then populates an SQLite database called the Integrated Proteomics database (IPdb), and makes the linked data accessible through (i) a local SQL interface, (ii) remote RESTful web service or (iii) a web browser.
