Figure 3.
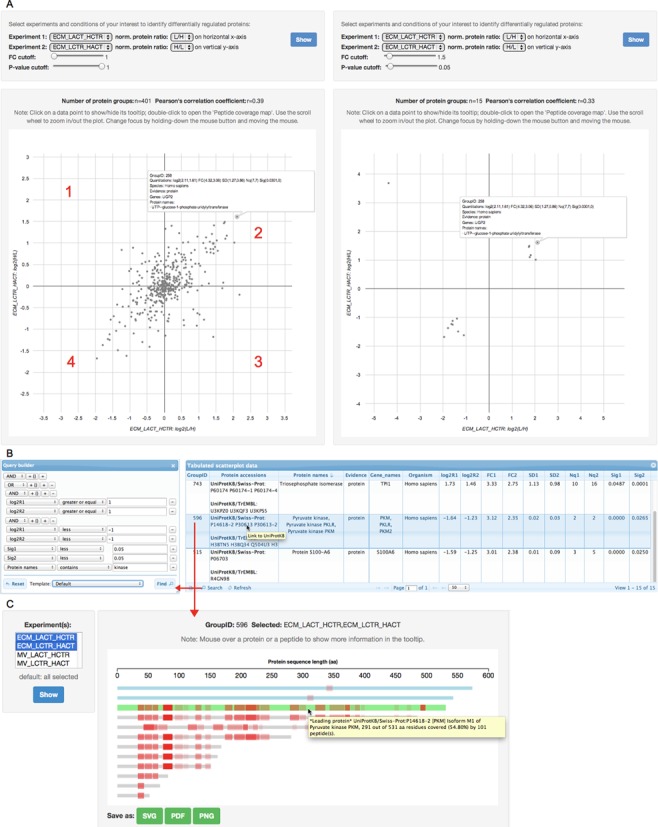
Interactive visualization and query tools available through the PIQMIe web interface. (A) 2D scatterplots of protein quantitations from reciprocal SILAC ECM experiments before (left figure) and after (right figure) the use of fold-change and intensity-based significance B cutoffs (FC ≥ 1.5; P value < 0.05). The plots are divided into four quadrants: the 1st and 3rd quadrants contain proteins that are inconsistently up- or down-regulated in the reciprocal experiments (false positives) whereas the 2nd and 4th quadrants contain proteins that are consistently up- and down-regulated by activin A signaling, respectively. For example, the UGP2 is consistently up-regulated in both ECM experiments. Moreover, the scatterplots are accompanied by the Pearson's correlation coefficient (r) computed for a pair of (reciprocal) SILAC experiments to aid in assessing the reproducibility of the replicate experiments. (B) Searchable protein grid with a query builder enables filtering of tabulated data by applying Boolean and/or relational operators on one or more columns of the grid. An example query is shown to select proteins with consistent (normalized) SILAC ratios from the set of potentially regulated proteins (FC ≥ 2; P value < 0.05) annotated as ‘kinase’. (C) Peptide coverage map shows the location and distribution of identified peptides (in red) within their parent proteins of a group. In the group (ID: 596), three protein entries belong to the manually curated UniProtKB/Swiss-Prot (in blue and green) including the ‘leading’ or best-scoring protein (accession: P14618-2, in green), as identified by the MaxQuant/Andromeda search, while the remaining nine proteins belong to the automatically annotated (unreviewed) UniProtKB/TrEMBL section (in gray). In addition, users can choose which experiments to view in the map. Note: All protein groups, accessions and peptides reported in the web interface are provided with hyperlinks to the appropriate site.
