Figure 1.
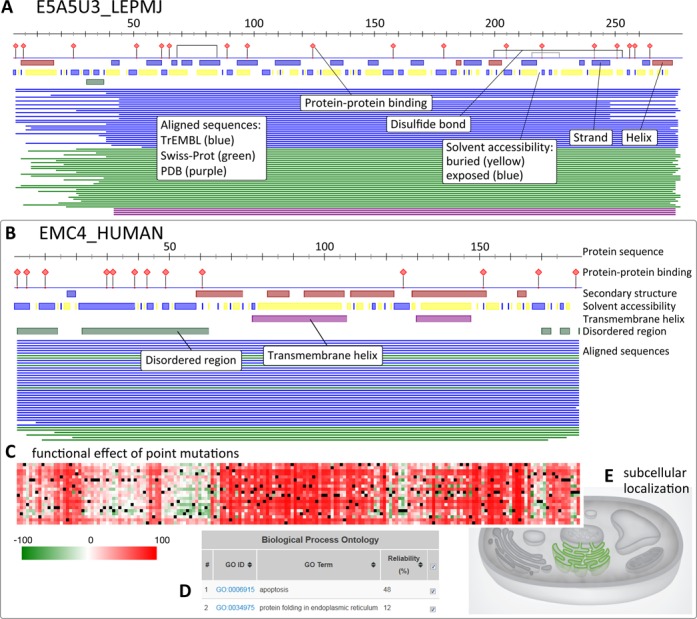
Visual results from PredictProtein (PP). The PP Dashboard Viewer shows a schematic of all position-based predictions and sequence alignments. (A) Putative protein (UniProt AC E5A5U3). (B) ER membrane protein complex subunit 4 (EMC4, UniProt AC Q5J8M3). The protein sequence is represented by a scale on top of the predicted features. Features presented include protein–protein binding sites (ISIS2), disulfide bonds (DISULFIND), structural features such as secondary structure state and solvent accessibility (PROFphd), transmembrane helices (TMSEG) and disordered regions (MD). Proteins aligned by PSI-BLAST (7) are shown as thin lines colored by database origin (PDB (11), Swiss-Prot (12) and TrEMBL (1)). Clicking on each line links to the database entry of the hit. For all elements, tooltips disclose the annotated feature, its position in the sequence and its type (prediction versus database search). (C) A complete analysis of the functional effect of point mutations on EMC4 shown in a heatmap (SNAP2). (D) Predicted GO terms (metastudent) for EMC4 in tabular format. (E) The predicted cellular compartment, ER membrane, for EMC4 (LocTree3) is highlighted in green in a schematic of a eukaryotic cell.
