Figure 1.
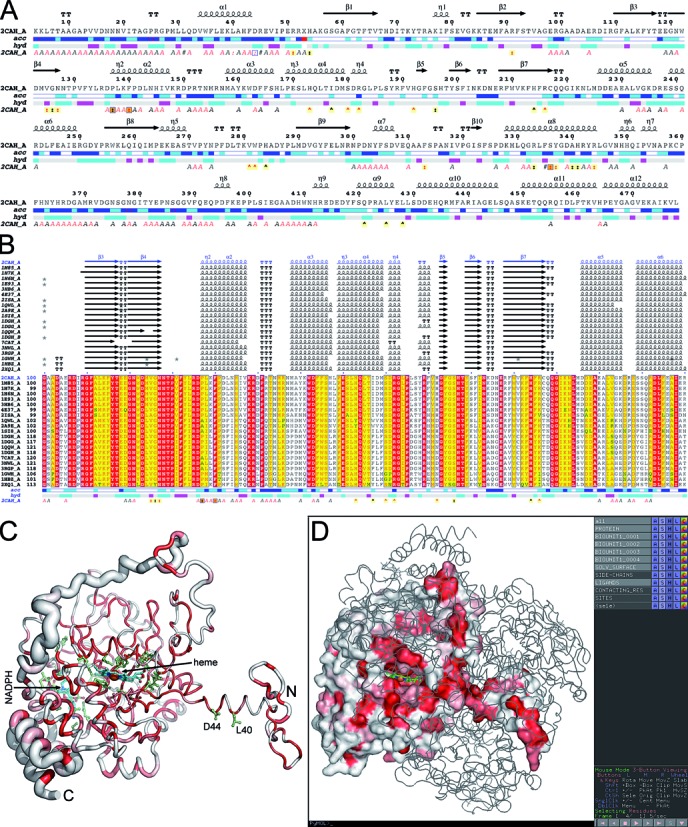
Sample of ENDscript representations obtained from the PDB entry 2CAH. (A) Flat figure showing the sequence of 2CAH with secondary structure elements presented on top (helices with squiggles, β-strands with arrows and turns with TT letters). Solvent accessibility is rendered by a first bar below the sequence (blue is accessible, cyan is intermediate, white is buried) and hydropathy by a second bar below (pink is hydrophobic, white is neutral, cyan is hydrophilic). Bottom letters and symbols depict crystallographic, protein:protein and protein:ligand contacts (see ‘Case study’). (B) Excerpt from the second ENDscript flat figure, showing the MSA of the 20 most homologous proteins to 2CAH (obtained with a BLAST+ search against the PDBAA database). Known secondary structure elements are displayed for all aligned sequences. Alternate residues are highlighted by gray stars. Identical and similar residues are boxed in red and yellow, respectively. (C) The ‘Sausage’ PyMOL representation showing a tube depiction of 2CAH, whose radius is proportional to the mean rms deviation per residue between Cα pairs. The tube is colored according to the level of sequence conservation, from white (low score) to red (identity). The heme cofactor and bound NADPH are presented in cyan ball-and-stick. Their contacting residues are presented in light green ball-and-stick. (D) Screen capture of a PyMOL session showing the biological tetrameric assembly of 2CAH. One monomer is presented by its solvent accessible surface with the same color ramping and orientation as in panel (C). The Cα traces of the three other monomers are presented. On the right, the PyMOL control panel with preset buttons allows to show and hide structural features compiled by ENDscript.
