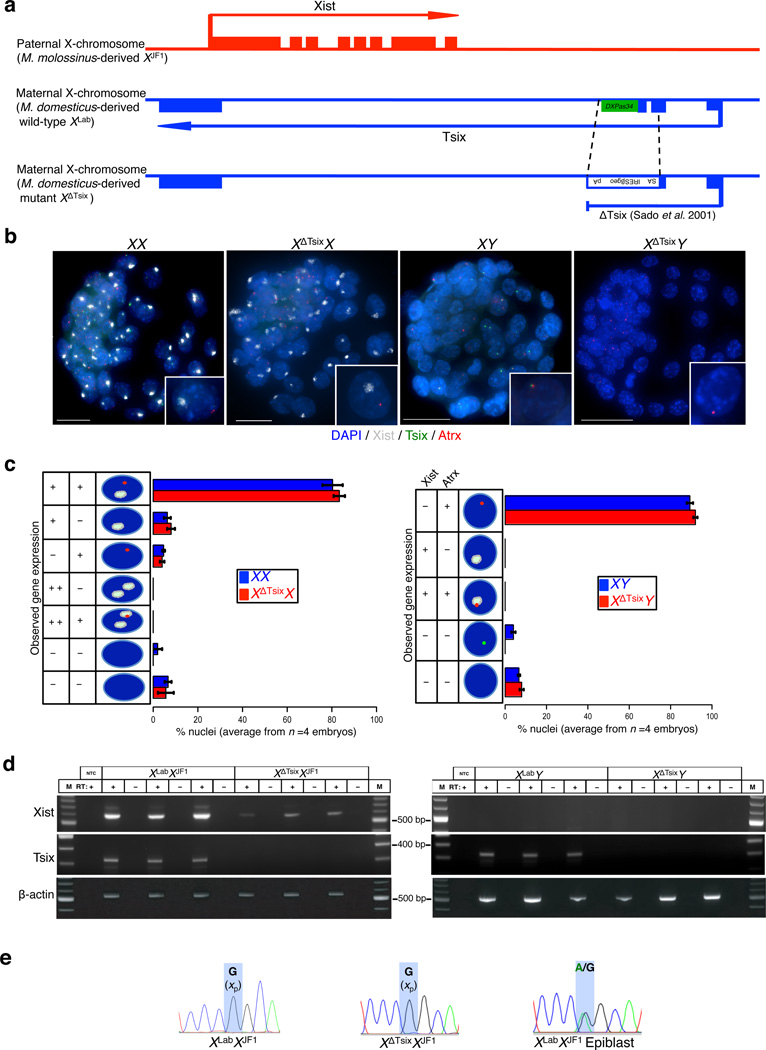
Figure 1. Absence of ectopic Xist induction from the XΔTsix maternal X-chromosome in embryonic day (E) 3.5 blastocyst embryos.
(a) Schematic representation of the genomic structure of Xist, Tsix, and the Tsix RNA truncation mutant XΔTsix(b) RNA FISH detection of Xist (white), Tsix (green) and Atrx (red) RNAs in representative E3.5 embryos. Nuclei are stained blue with DAPI. Insets show representative nuclei. Scale bar, 25 µm. (c) Quantification of Xist, Tsix, and Atrx RNA expression patterns in blastocyst nuclei. The X-axis of each graph represents the average % nuclei observed in each class for each genotype. n=4 embryos per genotype. Diagrams along the Y-axis depict all observed expression patterns. +, RNA expression detected from a single X-chromosome; + +, RNA expression detected from both X-chromosomes; -, absence of RNA detection. Gene expression pattern does not differ significantly between wild-type and Tsix mutant blastocysts (Fisher’s exact test). Error bars, S.D. (d) RT-PCR detection of Xist, Tsix, and control b-actin RNAs. Three individual embryos are shown for each genotype. M, marker; NTC, no template control; +, reaction with reverse transcriptase (RT); -, no RT control lane. (e) Sanger sequencing chromatograms of representative Xist RT-PCR products. Highlights mark a single nucleotide polymorphism that differs between the maternal XLab / XΔTsix alleles and the paternal XJF1 allele (see Methods). Both XLabXJF1 and XΔTsixXJF1 females express Xist only from the paternally-inherited X-chromosome (Xp). The XLabXJF1 epiblast is a control sample displaying expression from both parental alleles.