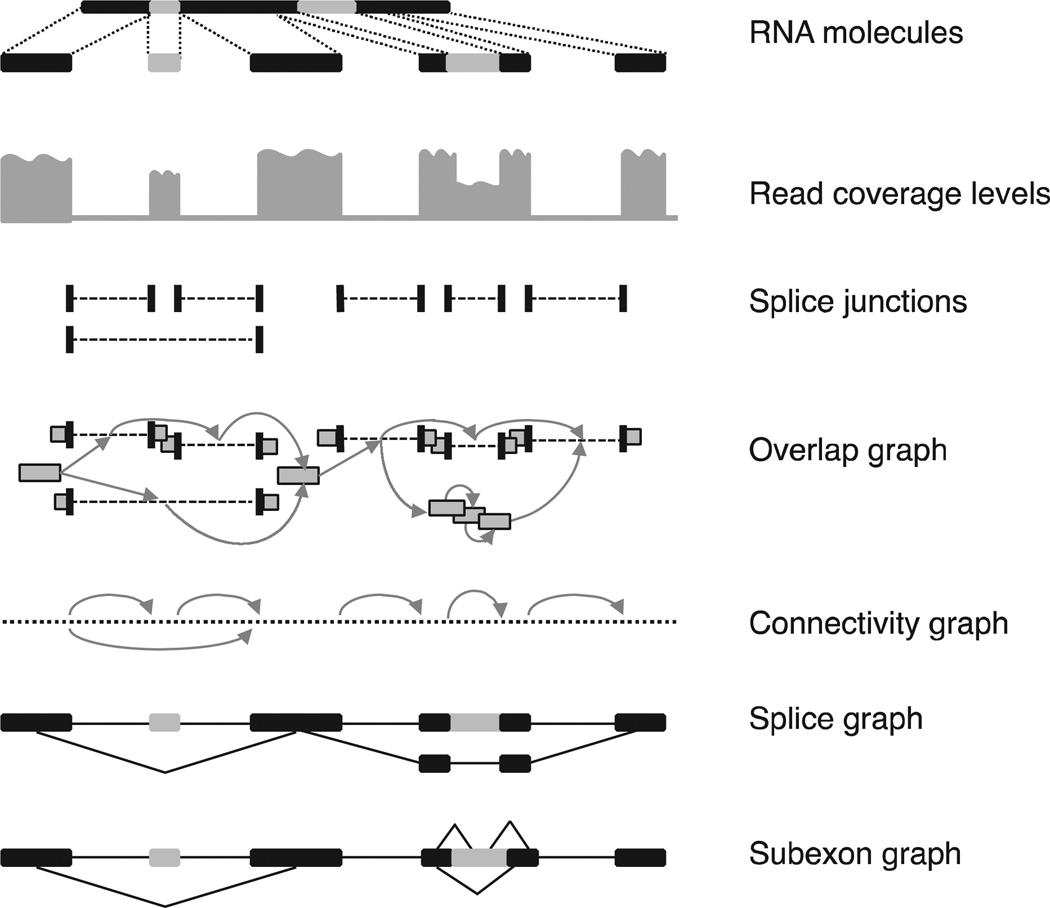
Fig. 2.
Overview of data structures employed by reference-based transcript assembly. (Top) RNA-seq molecules are sequenced and reads are aligned to the reference genome. Light-colored segments are alternatively spliced. Spliced reads define the introns of a gene, whereas exons are derived from both unspliced and spliced reads. Transcripts are assembled from the reads using various algorithmic techniques. An overlap graph has a node for each read, and two reads are connected by an edge iff they are compatible (i.e., they have the same splicing patterns along the overlap segment). A connectivity graph connects any two consecutive bases on the chromosome, as well as the endpoints of introns. For simplicity, only spliced edges are shown, with arrows. A splice graph has exons as nodes, connected by introns (edges); splice variants can then be read from the graph as maximal paths. As a variation, a subexon graph connects gene segments if they are adjacent on the genome as part of the same exon, or are connected via a spliced read.