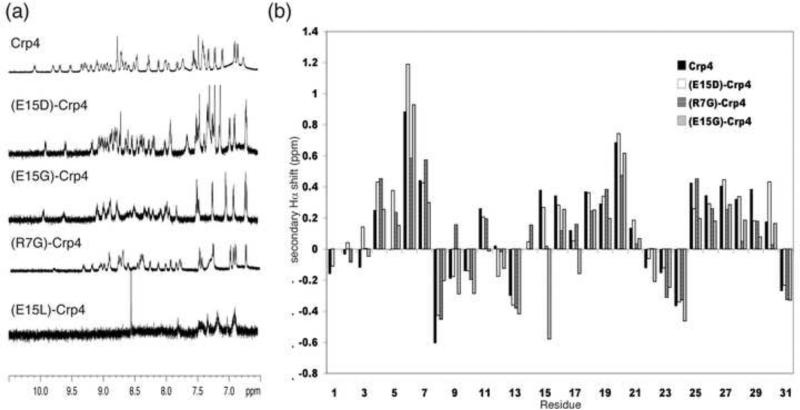
Figure 2.
NMR data on Crp4 and analogues. (a) 1D 1H spectrum of Crp4, (E15D)-Crp4, (E15G)-Crp4, (R7G)-Crp4 and (E15L)-Crp4. Native Crp4 as well as the (E15D)-, (E15G)- and (R7G)- analogues show good signal dispersion. In contrast (E15L)-Crp4 is misfolded and aggregates in solution, resulting in broad lines with amide signals not visible in the 1D experiment. (b) Secondary Hα chemical shifts for Crp4 and analogues. Similar trends with stretches of positive numbers characteristic of β-sheets are seen in all peptides, indicating that the native fold is retained despite the mutations to the salt-bridge.