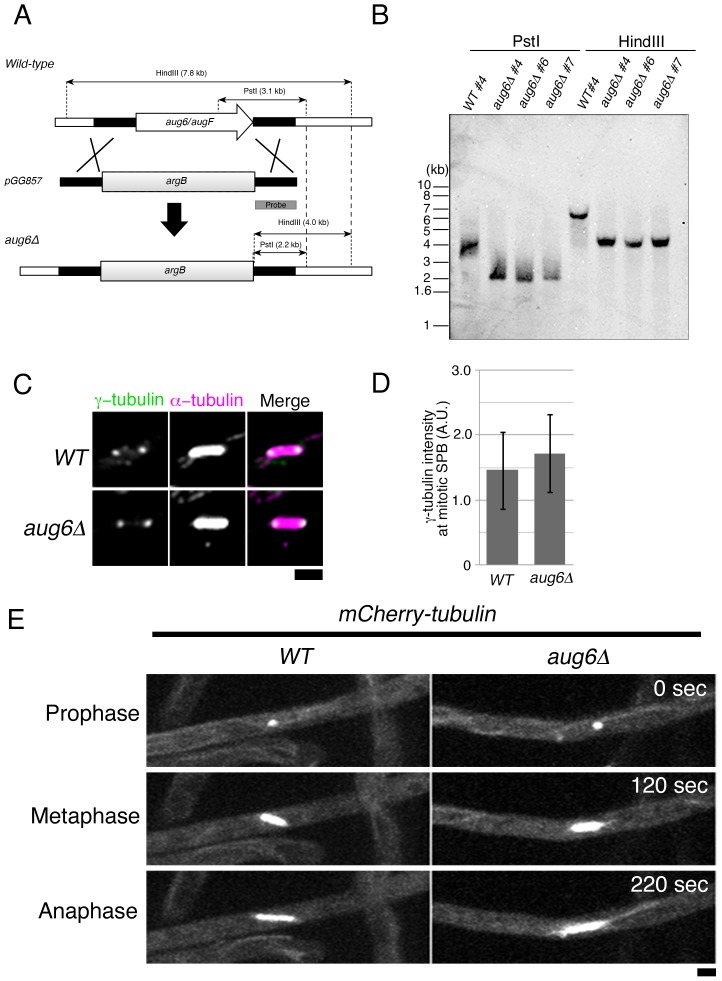
Figure 4. Normal γ-tubulin localisation and mitotic progression in the absence of augmin.
(A) Scheme showing the generation of the aug6 disruptant by one-step gene replacement. (B) Southern blot hybridisation confirmed the aug6 gene deletion via homologous recombination (3 independently selected strains were analysed). The probe is described in (A). We used the #4 strain in this study. (C, D) γ-tubulin localisation to the SPB was not attenuated in the absence of aug6. γ-tubulin signal intensities were quantified in the spindle of WT and aug6Δ in 2 independent experiments, and the result from one experiment is displayed (n = 9 hyphae, total 77 and 73 spindles, respectively). Error bars indicate SD. When multiple spindles were analysed in a hypha, the mean value was used as the γ-tubulin signal intensity of the hypha. Maximum projection images of 7 z-sections are displayed. (E) Normal spindle formation and mitotic progression in aug6Δ, as monitored by time-lapse imaging of mCherry-tubulin. 25 spindles of 10 WT cells and 22 spindles of 12 aug6Δ cells were analysed. Maximum projection images of 5 z-sections are displayed. See also Movie S3. Bar, 2 µm.