Figure 1.
A Structure Search on the Level of Multiprotein Complexes Reveals Significant Structural Correlations that Are Undetectable on the Level of Single Chains
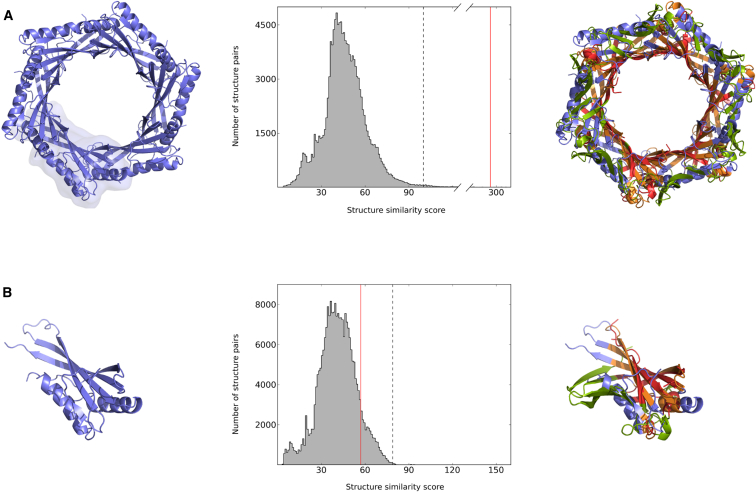
(A and B) In order to identify similarities to other protein structures, a cytoplasmic protein of unknown function from S. typhimurium (PDB ID code: 2GJV; to be published) is compared to all proteins in PDB. (A) Left: structure of homohexameric 2gjv@1, with chain A contoured. Middle: distribution of structure similarity scores obtained from a search of 2gjv@1 against all known biological assemblies (138,294 items; November 7, 2013). Extensive structure similarity (S = 292.2, red vertical line) is found between 2gjv@1 and homohexameric secretory protein Hcp3 from P. aeruginosa (3he1@1; Osipiuk et al., 2011). Note that a score of 292.2 is far above the threshold of S+ = 100.0 (dashed line; Experimental Procedures) and close to the end of the distribution’s right tail; only 17 hits have a score S > 292.2. Right: superposition of 2gjv@1 (blue) and 3he1@1 (green), with matching parts in orange (2gjv@1) and red (3he1@1). (B) Left: structure of chain A of 2gjv. Middle: distribution of structure similarity scores obtained from a search of chain A of 2gjv against all known protein chains (242,925 items; November 7, 2013). As indicated by the red vertical line, the structure similarity S = 56.7 to monomeric Hcp3 (chain A of 3he1) is hardly distinguishable from random matches and quite below the threshold of S+ = 79.0 (dashed line). More than 25,000 other protein chains yield a score S > 56.7 when compared to chain A of 2gjv. Right: superposition of chains A of 2gjv and 3he1, respectively. Colors as in (A).