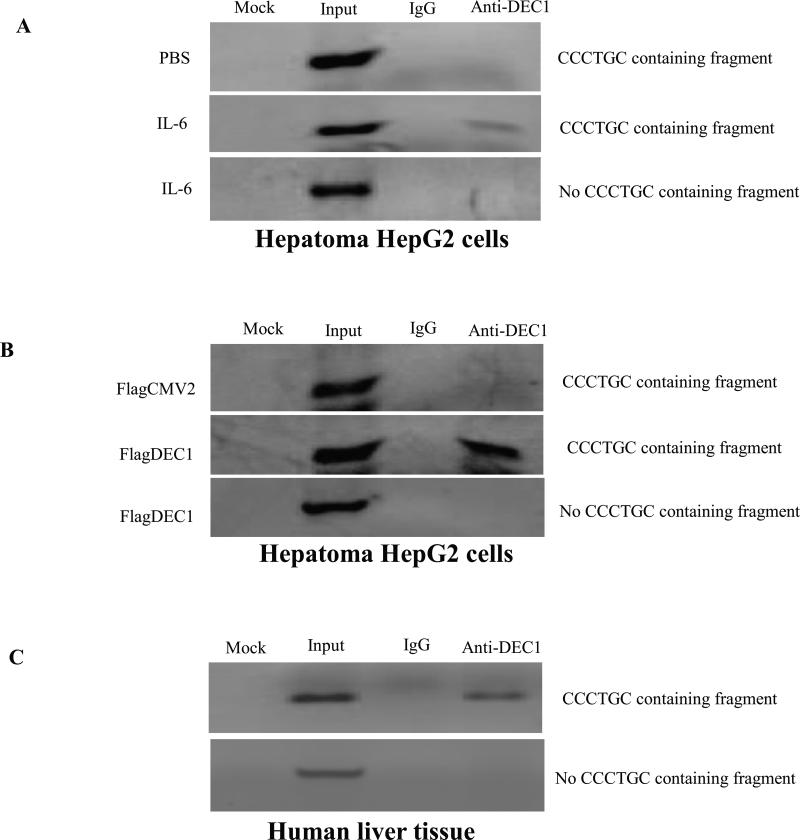
Fig.6. DEC1 being specifically recruited to the CCCTGC sequence in the CYP3A4 promoter by ChIP.
(A) HepG2 cells treated with IL-6. (B) HepG2 cells transfected with DEC1 expression construct. The chromatins with optimized size ranging from 400 to 800 bp were prepared as described in Materials and Methods. After immunoprecipitation with antibodies against DEC1 or IgG as a negative control, one set of PCR amplification was performed to specifically detect the presence of CCCTGC sequence-containing chromatin DNA. A negative control primer (no CCCTGC sequence containing fragment) set was included to amplify a 206 bp fragment 450 bp upstream of the CCCTGC sequence. PCR amplification was carried out in a final volume of 25 μl with 2 μl of eluted chromatin DNA, input DNA, or water (mock) as template with 40 cycles. PCR products were resolved on a 1.5% agarose gel. Three independent experiments were performed. (C) Chromatins were prepared from normal human liver tissues and immunoprecipitated with antibodies against DEC1 or IgG. The presence of CCCTGC sequence-containing chromatin DNA was detected by amplification using a set of primers flanking CCCTGC sequence. PCR amplification using the no CCCTGC sequence primers set was also performed as a control (n = 3).