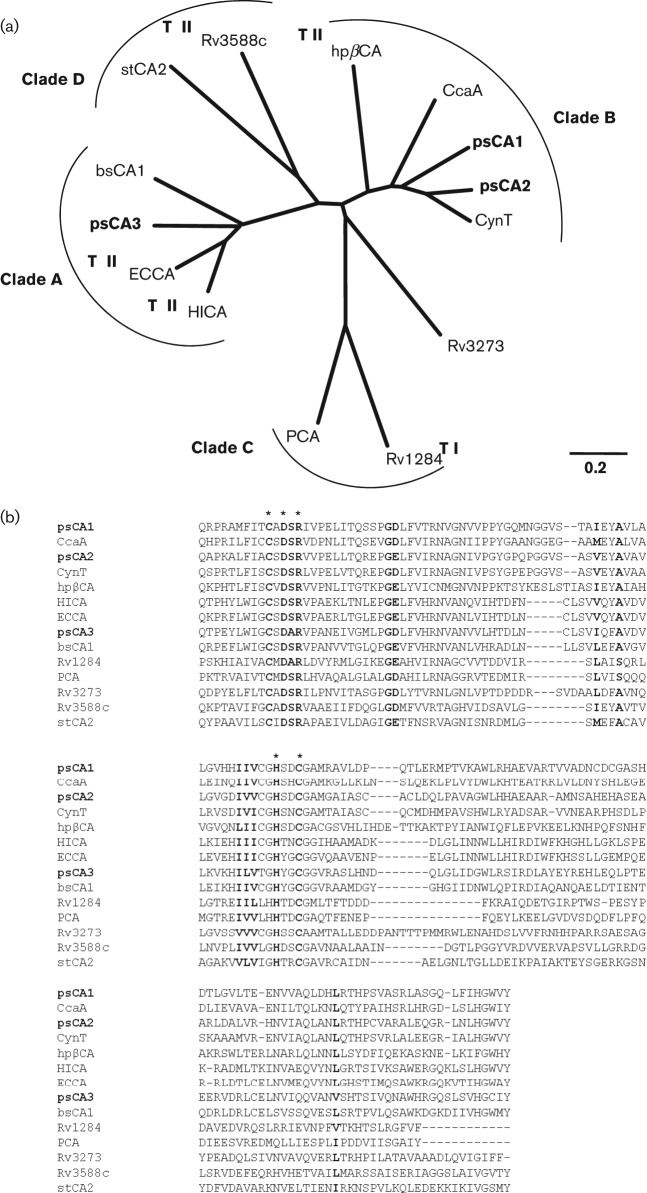
Fig. 1.
Sequence analyses of psCA1, psCA2 and psCA3. (a) Unrooted bootstrap neighbour-joining phylogenetic tree calculated from an alignment of the amino acid sequences of the PAO1 CAs and the following bacterial β-class CAs: gi|78099988| CynT E. coli, gi|2493490| CcaA Synecocystis, gi|6014888| hpβCA He. pylori, gi|47606320| ECCA E. coli, gi|1175500| HICA Ha. influenza, gi|23500522| bsCA 1 B. suis, gi|54042506| Rv1284 M. tuberculosis, gi|332075681| PCA St. pneumoniae, gi|15610409| Rv3273 M. tuberculosis, gi|15610724| Rv3588c M. tuberculosis, gi|16445318| stCA 2 Sa. Typhimurium. TI and TII represent type I and type II β-CAs, respectively. Bar, 0.2 changes per nucleotide position. (b) Amino acid sequence alignment of the predicted active sites of the PAO1 CAs and the eleven bacterial β-CAs used in the phylogenetic analyses. The active site residues of the CAs were predicted by using the CDD algorithms in the NCBI (Marchler-Bauer et al., 2011). The conserved amino acids are shown in bold. Five residues involved in Zn2+ coordination and catalytic activity are shown with an asterisk (Joseph et al., 2010).