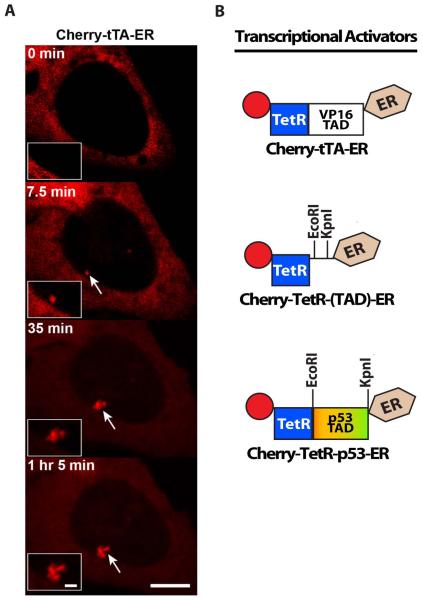
(B) Schematic diagram of the synthetic transcriptional activator constructs, which can be used to analyze the effects of different transcriptional activation domains (TADs) on transcription and chromatin organization in cell lines with arrays that include the tetracycline response element repeats (
Figure 1). Cherry-tTA-ER consists of the auto-fluorescent protein, Cherry, the tetracycline transcriptional activator (tTA), and the estrogen receptor hormone-binding domain (ER). tTA, itself, is a fusion between the tetracycline repressor (TetR) and the VP16 TAD. Cherry-TetR-(TAD)-ER was constructed for easy introduction of dirent TADs by directional cloning using EcoRI and KpnI. Cherry-TetR-p53 was constructed by introducing the p53 TAD into Cherry-TetR-(TAD)-ER.