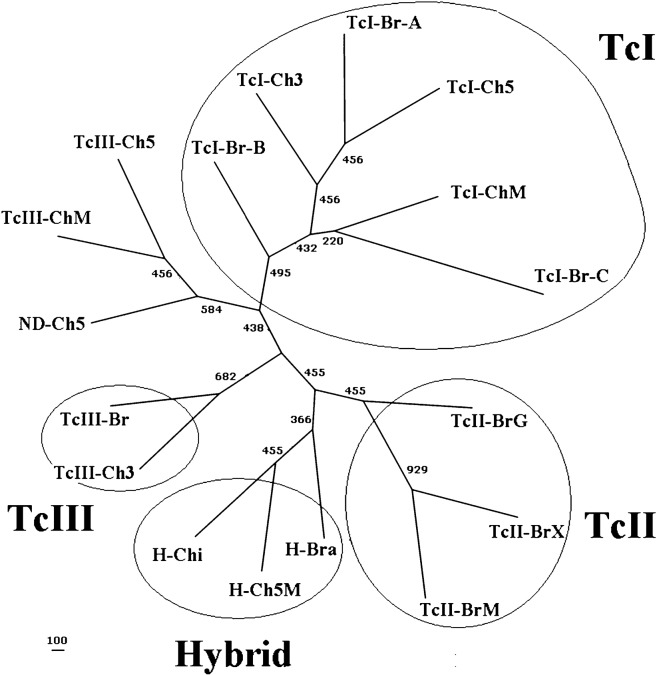
Fig. 4.
Unrooted phylogenetic tree of the T. cruzi population samples from the midgut of triatomines from Chilean and Brazilian regions. The phylogenetic reconstruction was performed using the allele sharing distance method (Bowcock et al., 1994) and the SCLE10, SCLE11 and MCLE01 microsatellite markers (Oliveira et al., 1998). The T. cruzi population samples from triatomine midgut were directly genotyped and correspond to groups of insects collected in the III (TcI-Ch3 and TcIII-Ch3), V (TcI-Ch5, TcIII-Ch5 and ND-Ch5) and Metropolitan (TcI-ChM and TcIII-ChM) Chilean regions. The lineage nomenclature corresponds to the recent agreement (Zingales et al., 2009); they were assigned based on three simultaneous methods and correspond to the following T. cruzi population samples: TcI lineage (TcI-Ch3, TcI-Ch5 and TcI-ChM) and TcIII lineage (TcIII-Ch3, TcIII-Ch5 and TcIII-ChM). H-Ch5M corresponds to a T. cruzi population sample of the TcV–TcVI lineage from triatomine midguts collected in the V and Metropolitan Chilean regions. The laboratory cultured T. cruzi population samples correspond to published data of de Freita et al. (2006) and Venegas et al. (2009a) and are the following: TcI from Brazil Amazon (TcI-Br-A), TcI mainly from Brazil-Minas Gerais (TcI-Br-B), TcI from Bolivia, Colombia and France Guyana (TcI-C), TcII from Brazil-Goias (TcII-Br-G), TcII from Brazil-Minas Gerais (TcII-Br-M), TcII from several Brazilian regions (TcII-Br-X), TcIII mainly from Minas Gerais (TcIII-Br), TcV–TcVI from Minas Gerais (H-Bra) and TcV–TcVI from Chile (H-Chi) (see Table 3 and the section on ‘Materials and methods’ for more details).