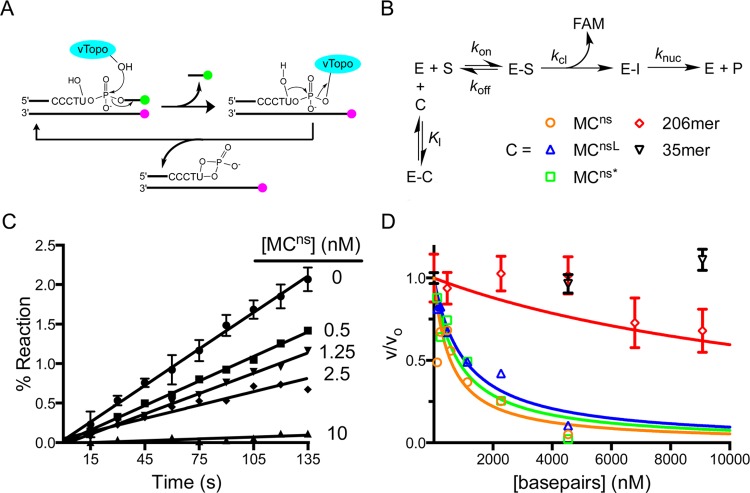
Figure 3.
Competition assay for measuring the binding affinity of vTopo for various topological isoforms of MCns and nonspecific linear DNAs of varying lengths. (A) Schematic of the continuous multiple-turnover fluorescence assay for measuring vTopo-catalyzed strand cleavage. The formation of a covalent complex between vTopo and the substrate releases a highly fluorescent 3′-FAM-labeled 7mer that is otherwise quenched in the duplex substrate by the IABK quench group on the opposite strand’s 5′-end. The presence of the uracil ribonucleotide in the cleavage sequence (CCCTU) results in the release of vTopo by nucleophilic attack by the 2′-OH group.29 Thus, multiple steady-state turnovers are possible, and initial linear rates can be accurately measured. (B) Competition binding mechanism. Because the Km for the fluorescent substrate is known, the Ki for each competitive inhibitor (C) can be determined from the concentration dependence of the decrease in fractional velocity (vi/v0) using eq 3. (C) Steady-state initial rates as a function of MCns concentration. Error bars have been removed for the sake of clarity but are similar in magnitude to those shown. The concentrations indicated are based on plasmid molecular weight. (D) Fractional velocity as a function of nucleic acid inhibitor concentration. To normalize for the different lengths of DNA used, the concentration is noted in molar base pairs. Most error bars have been omitted for the sake of clarity but are similar to those shown.