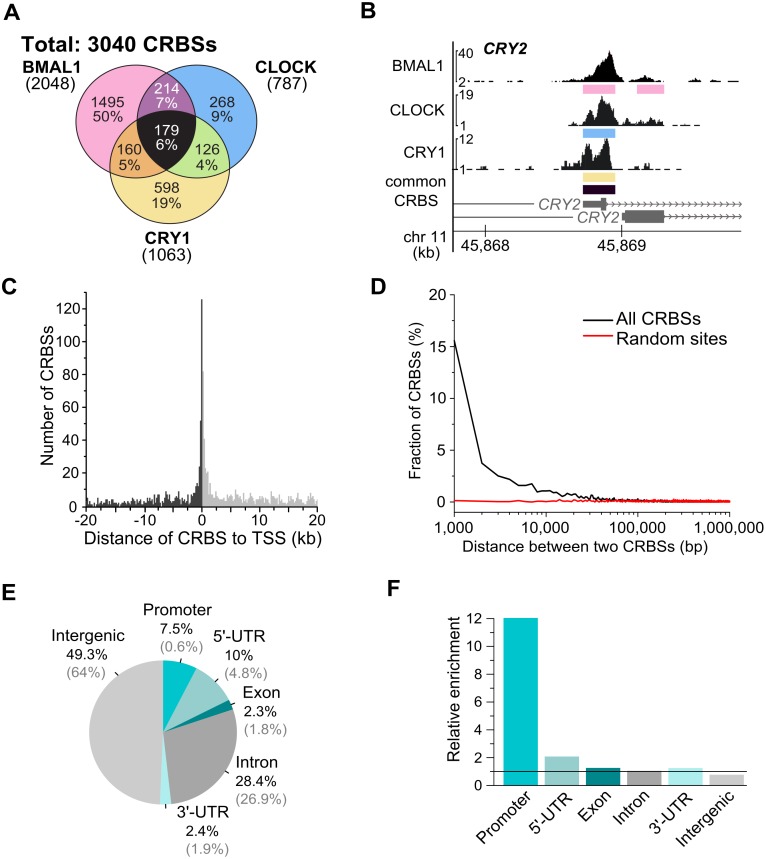
Figure 1. Genome-wide binding sites of the circadian transcription regulators BMAL1 CLOCK and CRY1.
(A) Venn diagram showing numbers and percentage of individual and overlapping binding sites of BMAL1, CLOCK, and CRY1. Percentages represent the fraction of the CRBSs over the total number of sites for the 3 proteins (B) UCSC browser views of BMAL1 (top), CLOCK (middle) and CRY1 (bottom) occupancy at the promoter of CRY2. Regions detected as binding sites of the individual transcription regulators are indicated by colored bars. Black bars indicate common binding sites of the three regulators. (C) CRBSs cluster near TSSs. Histograms of positions of all CRBSs are shown for a window of ±20 kb around TSSs with a bin size of 200 bp. 352 CRBSs are enriched ±1 kb around TSS. (D) Histogram of the distance between two consecutive CRBSs (black) in comparison to a set of 3040 random sites (red). (E) Pie chart showing the percentage of binding sites in a genomic region (black) and the contribution of the region (%) to the genome (grey). (F) Genomic annotation of the CRBSs to promoter (−1 kb to TSS), 5′UTR, exon, intron, 3′UTR, and intergenic region.