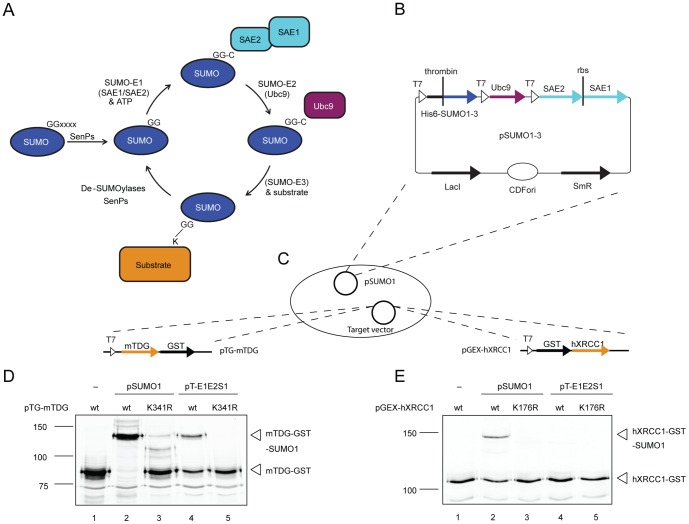
Figure 1. The pSUMO-based SUMOylation system modifies proteins in E.coli.
(A) Scheme of the in vivo SUMO maturation, SUMO conjugation and deconjugation process (for detailed description, see “Introduction”). (B) pSUMO vectors containing the humanized SUMOylation system consisting of N-terminally His6-tagged SUMO1, 2 or 3, the SUMO-conjugating enzyme E2 (Ubc9) and both subunits of the SUMO-activating enzyme (SAE1 and SAE2) as a cistronic expression unit with an internal ribosomal binding site (rbs). Expression of the respective cDNAs is under the control of a lac-repressor (LacI) regulated T7 promoter. (C) Scheme of the experimental setup of the pSUMO-based in-cell SUMO conjugation. E.coli BL21 cells were used containing pSUMO1 in combination with pTG-mTDG or pGEX-hXRCC1 plasmids were used for the co-expression of the complete SUMO system with C- and N-terminally GST-tagged mTDG and hXRCC1, respectively. Immunoblot analyses of mTDG (D) and hXRCC1 (E) SUMOylation in E.coli cells, expressing the SUMO target and the SUMO system from either the pSUMO1 or the pT-E1E2S1 plasmid (250 µM IPTG at 25°C for 2 h). Co-expression of target proteins mutated in the SUMO acceptor sites of mTDG (K341R) and hXRCC1 (K176R) were included to assess the specificity of the SUMOylation system.