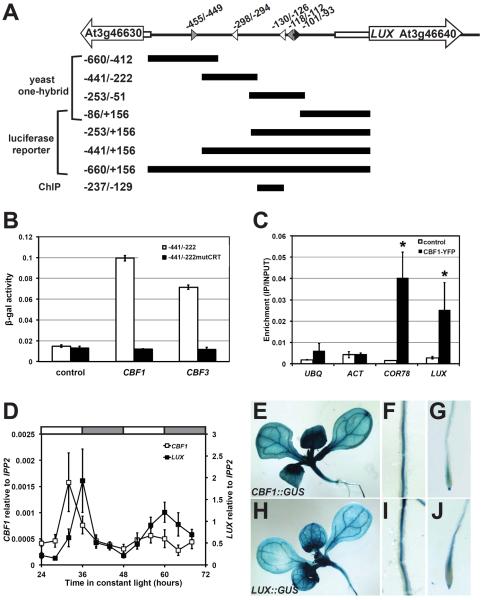
Figure 1.
CBF1 binds the LUX promoter
(A) Schematic of the LUX promoter. The transcriptional start occurs at +1. White arrows: gene bodies; white rectangles: 5'UTRs; arrowheads: DNA motifs with EELs in grey, CRTs in white, and EE in black; and black bars: promoter regions tested in different assays. Positions and sizes are roughly drawn to scale. (B) CBF1 binding to LUX promoter in yeast. Bars represent β-galactosidase activity where error bars represent standard error of three replicates. (C) CBF1 binding to the LUX promoter in Arabidopsis. ChIP assays were performed using control (LUX∷LUC −660/+156 line 139) or CBF1 overexpressing line 2–1 (CBF1ox2-1). Plants were grown under 12:12 and collected 16 hours after lights on for ChIP using an anti-GFP antibody. Results are normalized to input DNA. Bars represent average quantification from real-time PCR with error bars representing standard error of two independent experiments. Student's t-test was used to determine the significance of target enrichment relative to control (* p-value ≤0.05). UBQ, UBIQUITIN; and ACT, ACTIN. (D) Temporal expression of CBF1 and LUX. Wildtype plants were entrained under 12:12 before release to constant light at 22°C. mRNA levels were quantified by real-time reverse transcription PCR and normalized to IPP2. Data is the average of three biological replicates with error bars representing standard error. (E–J) Spatial expression of CBF1 and LUX. Seedlings carrying promoter∷GUS constructs of CBF1 (E–G) and LUX (H–J) were grown under 12:12 at 22°C. Expression detected throughout cotyledons, rosette leaves and hypocotyl (E and H); roots (F and I); and root tips (G and J).