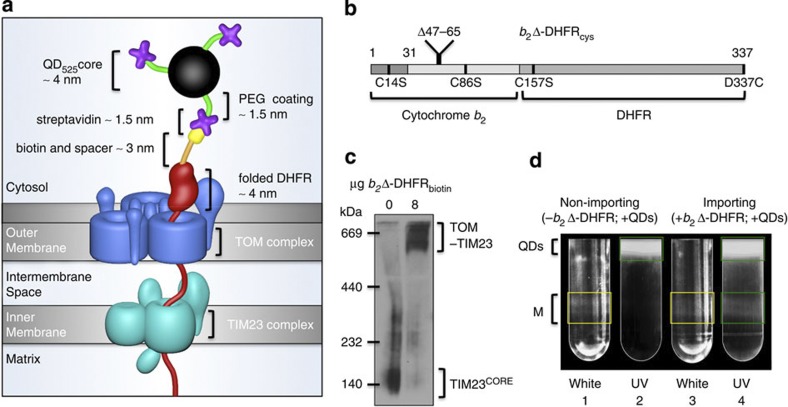
Figure 1. Design and implementation of the approach.
(a) Schematic of the STAMP (Specifically TArgeted Membrane nanoParticle) approach. In the first step, a small biotin label (yellow) is attached via a spacer arm (orange) to a mitochondrial model preprotein (red). Subsequently, the labelled preprotein is targeted to the mitochondrial import machinery and arrested by means of a tightly folded DHFR domain as a translocation intermediate spanning both TOM (blue) and TIM23 (cyan) complexes. In the third step, the DHFR-linked biotin is bound by streptavidin (purple)-conjugated QDs (black sphere with green spacer arms). The total distance from the QD to the outer mitochondrial membrane is ~10 nm. (b) The design of the mitochondrial model preprotein is based on the fusion protein b2Δ-DHFR18. To enable site-specific biotinylation, endogenous cysteines (C14, C86 and C157) were substituted with serine and a unique cysteine replaced the C-terminal residue D337. (c) Blue native electrophoresis and western blot analysis of the preprotein-tethered TOM–TIM23 supercomplex detected with an antibody against Tim23. Note that the TIM23 core complexes (TIM23CORE) are quantitatively shifted into TOM–TIM23 supercomplexes on addition of b2Δ-DHFRbiotin, indicating that the import sites are occupied by preproteins under these conditions. The experiment was repeated three times. (d) Free QDs were separated from labelled mitochondria on an OptiPrep gradient. Under white light (tubes 1 and 3) mitochondrial membranes are visible (M, yellow boxes), and under UV excitation, QD525 is detected (tubes 2 and 4; green boxes). QD525 co-localization with mitochondria is seen in tube 4.