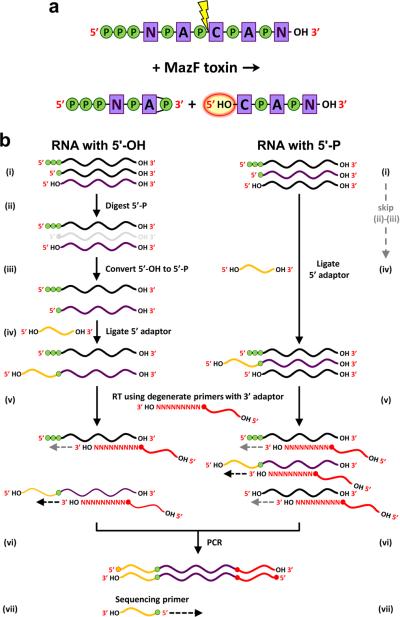
Figure 1. MORE RNA-seq cDNA library construction.
(a) Cleavage of RNA with MazF toxins yields two fragments – one with a 2’,3’-cyclic phosphate and one with a 5’-hydroxyl. P, phosphate; N, A, C, nucleosides; OH, hydroxyl. (b) Procedures for generating cDNA libraries derived from RNAs carrying only a 5’ hydroxyl (5’-OH) or only a 5’ monophosphate (5’-P). (i) Depicted are RNAs with a 5’ triphosphate, a 5’-P, or a 5’-OH; the species that will be converted into cDNA is shown in purple, and phosphate groups are depicted by green circles. Steps (ii) and (iii) are specific to generation of cDNAs from 5’-OH. (ii) RNAs with a 5’-P are removed by treatment with a 5’-P-specific exonuclease. (iii) RNA with a 5’-OH is phosphorylated by a kinase. (iv) The 5’ adaptor is ligated to RNAs that carry a 5’-P. (v) Reverse transcription (RT) using a primer with a 9-nt degenerate sequence at the 3’ end and the sequence of the 3’ adaptor at the 5’ end. (vi) PCR amplification using primers that match the 5’ and 3’ adaptors (only cDNAs that contain both 5’ and 3’ adaptor sequences can be amplified). (vii) The SOLiD sequencing ligation reaction originates from a primer complementary to the 5’ adaptor. The first base of each sequencing read corresponds to the 5’ end of an RNA transcript.