Figure 3. MazF-mt3 cleaves several M. tuberculosis mRNAs.
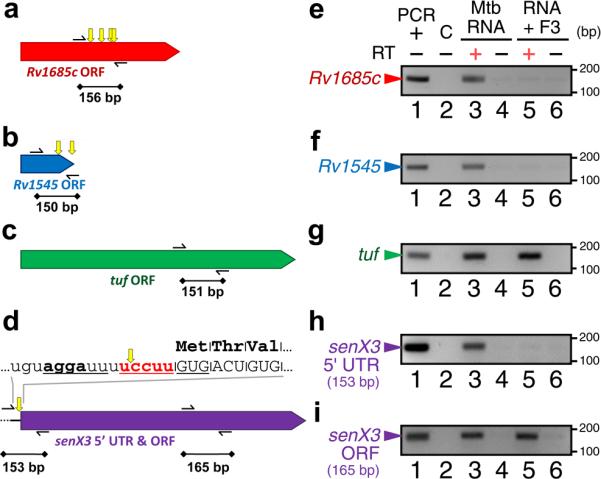
Scale schematics of M. tuberculosis genes that contain either two or more MazF-mt3 UCCUU motifs (a, b), no MazF-mt3 motifs (c), or one UCCUU motif in the 5’ UTR but none in the ORF (d). Thin black arrows correspond to primers used for RT-PCR, while black lines with diamonds correspond to regions amplified by RT-PCR. Yellow arrows above genes show the location of UCCUU motifs. (d) Text above the schematic represents the RNA sequence of the senX3 5’ UTR (lowercase) and ORF (uppercase), with the lone MazF-mt3 motif in bold red text and a putative SD sequence in bold text. (e-i) RT-PCR of M. tuberculosis total RNA incubated with (lanes 5 and 6) or without (lanes 3 and 4) purified MazF-mt3 and with (lanes 3 and 5) or without reverse transcriptase (RT; lanes 1, 2, 4, and 6). ”PCR +” indicates amplification using the appropriate PCR fragment as a template (lane 1), while ”C” is a negative control with H2O as a PCR template (lane 2). Length of DNA size marker in bp is noted by tick marks on the right. After treatment with MazF-mt3, regions that contain one or more UCCUU motifs (e, f, and h) no longer generated amplified products. This MazF-mt3-mediated cleavage was specific to regions containing one or more UCCUU motifs, since the amplified products in tuf (g) and in the senX3 ORF (i) were comparable in the absence (lane 3) or presence (lane 5) of MazF-mt3. Data shown are representative of two independent experiments. Complete gel images for (e-i) are shown in Supplementary Fig. 2.
