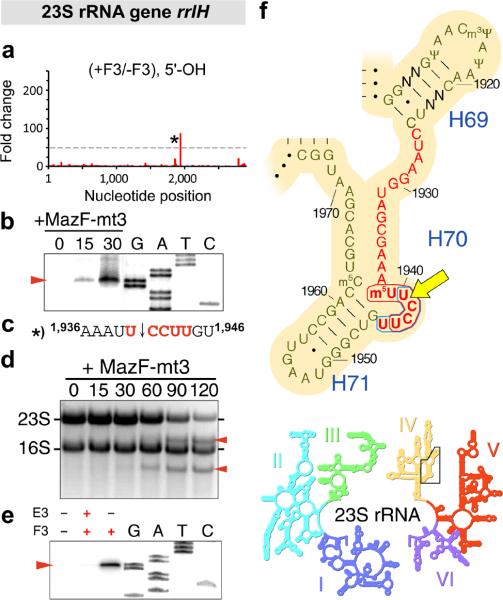
Figure 4. MazF-mt3 cleaves 23S rRNA at a conserved UCCUU within helix/loop 70.
(a) Histogram representing the ratio of #5’-ends observed in the analysis of 5’-OH from cells that did contain (+F3) or did not contain (-F3) MazF-mt3 within the E. coli 23S rRNA gene rrlH. The lone cleavage site is labeled with an asterisk (*). (b) Primer extension analysis of 23S rRNA isolated from E. coli cells at the indicated time (0, 15, and 30 min) after the induction of MazF-mt3 expression. “G, A, T, C,” sequencing ladder. The red arrow indicates the position of a cleavage product. (c) The RNA sequence surrounding the cleavage site (indicated by a black arrow) identified by MORE RNA-seq (a) and primer extension analysis (b), with flanking position numbers from E. coli 23S rRNA. (d) Ethidium bromide staining of total RNA isolated from E. coli cells at the indicated time (in min) after the induction of MazF-mt3 expression. (e) Primer extension analysis of 23S rRNA upon incubation of MazF-mt3 (F3) or MazE-mt3 (E3) with M. tuberculosis total RNA. Complete gel images for (b, d, e) are shown in Supplementary Fig. 4. (f) Sequence and secondary structure of a conserved region of domain IV (above) in 23S rRNA (inset below). The nucleotides that comprise helix/loop 70 (H70) are highlighted in red text. Bases that are not conserved between E. coli and M. tuberculosis are labeled “N,” E. coli position numbers are adjacent to tick marks, the MazF-mt3 recognition sequence is encircled by a blue line, the MazF-mt6 recognition sequence is encircled by a red line, and the site cleaved by both MazF-mt3 and MazF-mt6 is indicated by a yellow arrow. I-VI, numbered domains of 23S rRNA. Images in panel (f) are adapted with permission from http://rna.ucsc.edu/rnacenter/ribosome_images.html , Center for Molecular Biology of RNA, University of California, Santa Cruz.