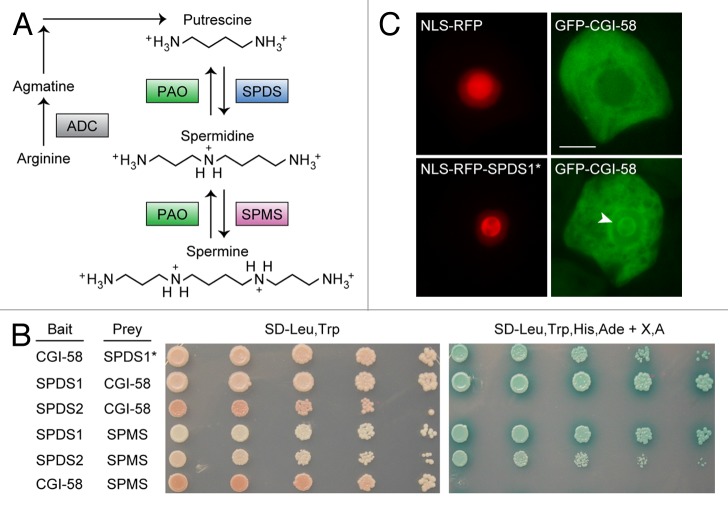
Figure 1. Interaction of CGI-58 and SPDS1. (A) Model of polyamine metabolism in plants (adapted from ref. 12). Key enzymes are highlighted in colored boxes, some of which are encoded by multiple genes. See text for additional details. Abbreviations include ADC, arginine decarboxylase; PAO, polyamine oxidase; SPDS, spermidine synthase; and SPMS, spermine synthase. (B) Yeast 2-hybrid analysis of protein-protein interactions between CGI-58 and various polyamine-related proteins, namely SPDS1, SPDS2 and SPMS. The indicated “bait” and “prey” proteins were co-expressed in yeast cells then similarly prepared serial (1:5) dilutions of yeast cultures were replica plated on either synthetic dextrose media lacking leucine and tryptophan (SD-Leu-Trp), which selects only for the presence of both “bait” and “prey” plasmids in the cells, or on media also lacking histidine, adenine, and containing X-α-Gal and Aureobasidin A (SD-Leu,Trp,His,Ade + X,A), which selects for 4 different reporter genes whose transcriptional activity is dependent on 2-hybrid protein interaction, as previously described.6 Note that the SPDS1* “prey” protein (top row) consists of only the C-terminal portion of SPDS1 (amino acids 131–334), whereas all other indicated “prey” (and “bait”) proteins are full length. (C) Protein-protein interaction analysis using the nuclear relocalization assay. Briefly, tobacco suspension-cultured (Bright Yellow-2) cells were transiently co-transformed (via biolistic bombardment; refer to ref. 6 for details) with plasmids expressing NLS-RFP and GFP-CGI-58 (upper panels), or a second set of plasmids expressing NLS-RFP N-terminally fused to the C-terminal half of SPDS1 (NLS-RFP-SPDS1*) and GFP-CGI-58 (bottom panels). Note in the bottom panels that a portion of the GFP-CGI-58 protein has been recruited from the cytosol into the nucleolus (refer to arrowhead) via its interaction with NLS-RFP-SPDS1*. Bar = 10 µm.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.