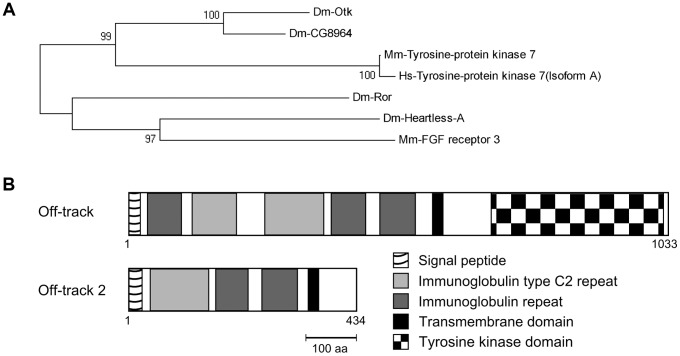
Figure 1. Off-track (Otk) and Off-track2 (CG8964, Otk2) are paralogs evolved by gene duplication.
(A) Phylogenetic tree representation of an alignment of sequences from different species homologous to Drosophila Otk. Blast-P of Dm-Otk in Drosophila returned Dm-CG8964 (Otk2) as the best hit; Dm-Ror and Dm-Heartless-A were the second and third hit, repectively. Blast-P of Dm-Otk in Mouse returned Mm-PTK7 as the best hit, while Mm-FGF receptor3 was the second best hit. Blast-P of Dm-Otk in Human returned Hs-PTK7 as the best hit. ClustalW alignment of these sequences and Neighbor-Joining tree confirms that the PTK7 branch is separated from the other proteins by a bootstrap value of 100. In this branch, two Drosophila genes, but only one in the other species can be found. (B) Protein structures of Otk and Otk2. Domains were predicted using the SMART sequence analysis tool. Dm, Drosophila melanogaster; Mm, Mus musculus; Hs, Homo sapiens.