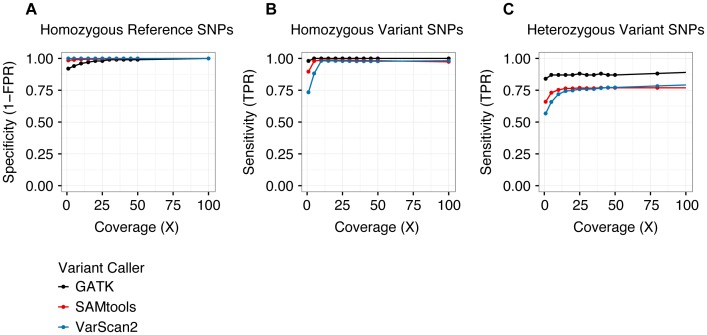
Figure 2. Performance of variant calling.
The specificity (A) and sensitivity (B, C) of three separate variant callers—SAMtools, VarScan2, and GATK—were evaluated by analyzing single-cell variant calls at germline SNPs previously ascertained by Affymetrix SNP arrays [14]. As we have defined true positive and true negative, sensitivity is undefined at homozygous reference positions (there are no true positives) and specificity is undefined at heterozygous and homozygous variant positions (there are no true negatives). Sensitivity and specificity were similar among all three callers at homozygous positions, but GATK demonstrated greater sensitivity at heterozygous sites. Variants were called jointly across all single-cell libraries with the GATK Unified Genotyper utility, whereas variants were called independently for each sample using SAMtools and VarScan2. See Table 2 and Table S6 for additional details. TPR: true positive rate. FPR: false positive rate.