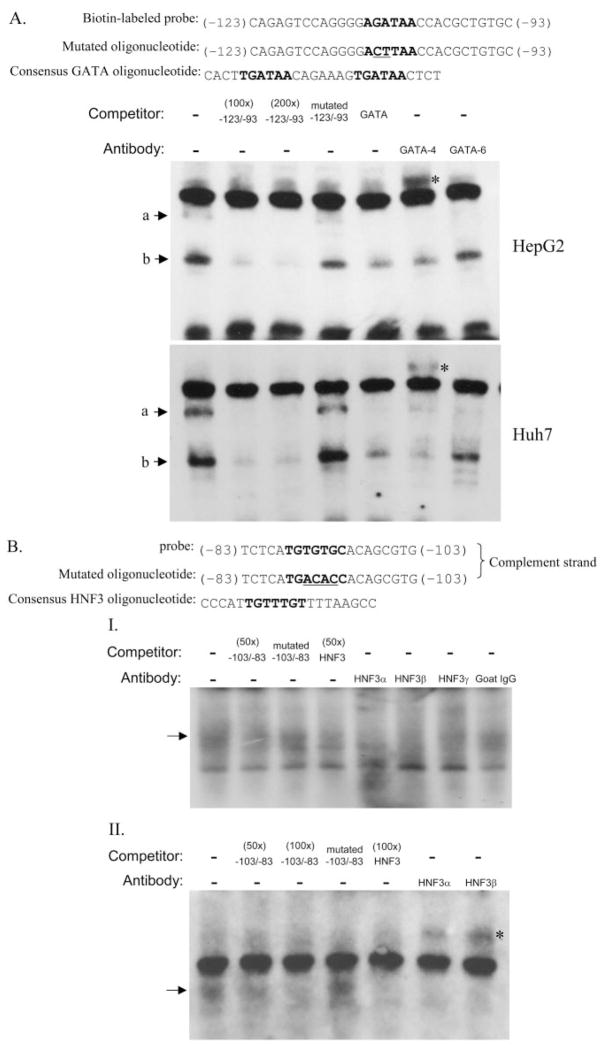
Fig. 8.
Gel mobility shift assay for GATA and HNF3 binding sites. A, competition and super-shift assays for the E1-GATA binding site using biotin-labeled GATA probe and nuclear extracts from either HepG2 or Huh7 cells. B, competitive and supershift EMSAs assays were conducted with using HepG2 nuclear extracts and E1-HNF3 DNA targets labeled either with radioactive (I) or biotin (II) methods. Labeled probes were mixed with different unlabeled competitors including GATA or HNF3 site-mutated oligonucleotides and consensus binding oligonucleotides whose sequences are shown on the top of each gel. The core binding sequences are shown in boldface letters, and the mutated bases are underlined. Two micrograms of the respective antibodies was added to the reaction mixture for supershift analysis. ☆, supershift complexes.