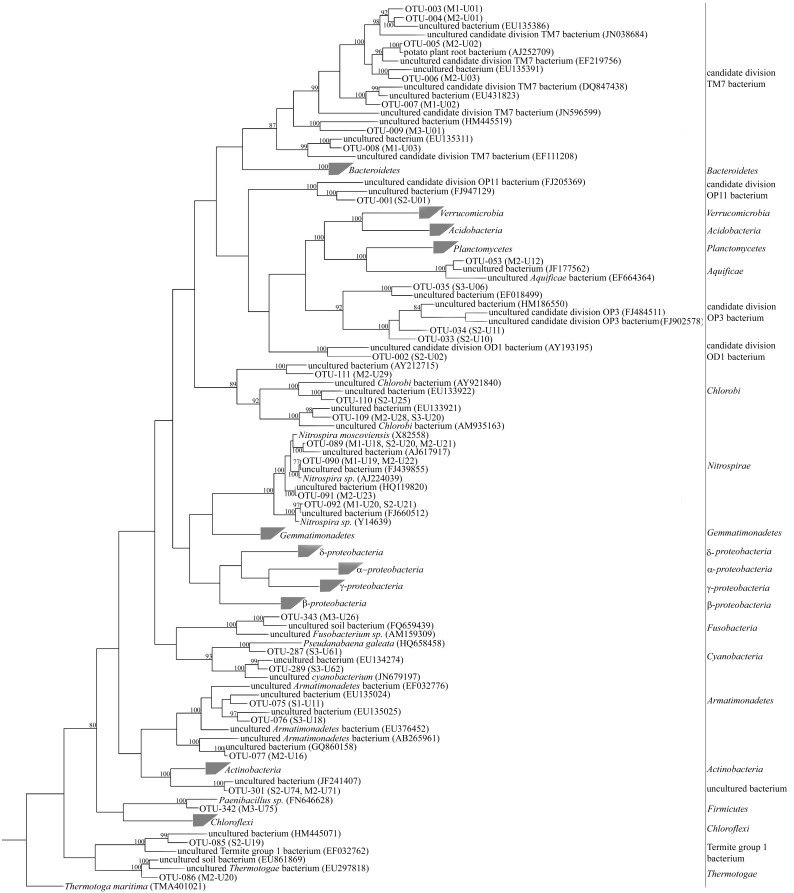
Figure 3. Phylogenetic tree of the bacterial sequences recovered from the six libraries, constructed using the neighbor-joining method with the Kimura two-parameter model for nucleotide change.
The predominant groups which were just denoted the phylogenetic positions in this tree, presented in other separate phylogenetic trees. The libraries OTUs occurred were labeled. Scale bar, 0.1 substitutions per nucleotide position. Bootstrap values (100 replicates) above 70% are indicated at the nodes. The tree was rooted using the sequence related to Thermotoga maritima (TMA401021).