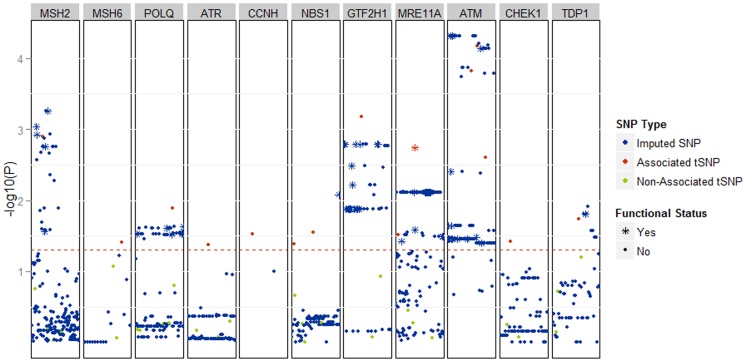
Figure 1. The results of association analysis, displayed as Manhattan plot, after imputation of 28 SNPs genotyped in both stage 1 and 2, which tag 11 DNA repair genes that showed association with NHL risk in our study.
The SNPs and genes are ordered by chromosomal position (x-axis). The associations are displayed as –log10(p-value) for each SNP. Red dots represent fifteen tagging SNPs that were genotyped in our study and were associated with NHL risk. Green dots represent tagging SNPs that were genotyped in our study and that showed no association with NHL. Blue markers represent SNPs imputed by IMPUTE from 1KG. The red dotted line defines the threshold of p-value <0.05. * indicates an associated SNP with a putatively functional impact; non-synonymous coding change or SNP mapping in: transcription factor binding site, H3K4Me1 chromatin mark, DNaseI hypersensitivity cluster, 5′UTR, 3′UTR.