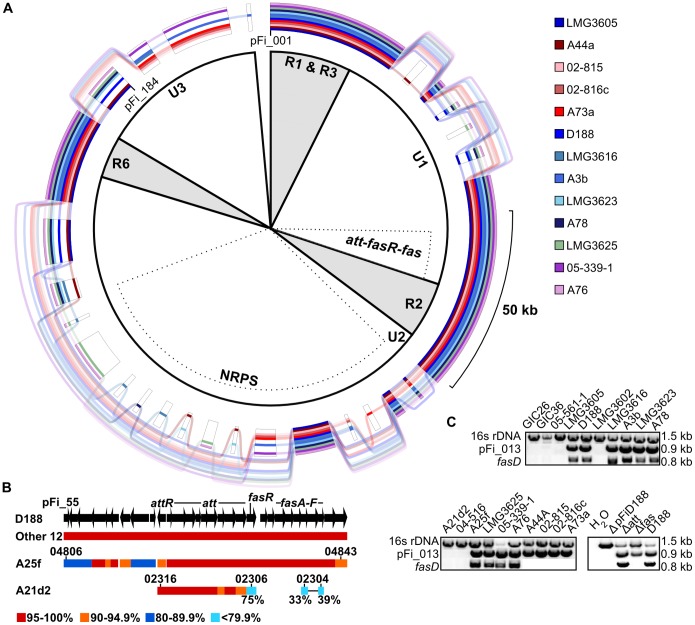
Figure 3. The att, fasR, and fas virulence loci are variable in organization among phytopathogenic isolates of Rhodococcus.
(A) GenomeRing of the 13 pFiD188-like sequences. Colored lines, corresponding to each isolate, indicate presence of a block. Transparent lines skip over absent blocks and connect co-linear blocks. A minimum block size of 1 kb was used. The inner portion defines conserved (R) and unique (U) regions of pFiD188 from isolate D188, as previously reported. The att-fas virulence loci and the NRPS-encoding region are also indicated by dotted regions. (B) Homology and co-linearity of the pFi_55-pFi_84 regions in the 15 phytopathogenic isolates. Top: structure of the region of pFiD188 from pFi_55 to pFi_84, with arrows depicting CDSs and the direction indicating the strand in which they are encoded. For isolates with a linear plasmid (Other 12), A25f, and A21d2, bars indicate the presence and range of sequence similarity. Relevant locus ID numbers are shown without RF25f_ and RFA21d2_ tags. The two cyan bars connected by a single line represent a gene fusion. (C) PCR-based detection of virulence loci (fasD) and pFiD188-like (pFi-013) sequences. The 16s rDNA gene was used as a positive control. Products were resolved on a 1%, 1X TAE agarose gel. Estimated product sizes (kb) are listed along the side.