Fig. 2.
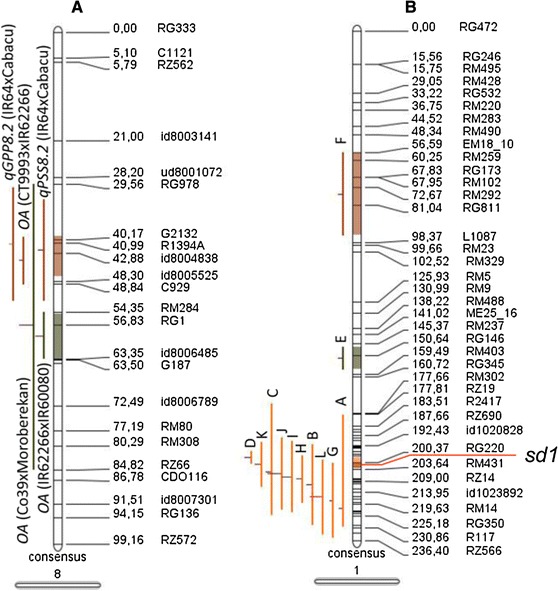
Genomic regions of rice chromosomes showing co-localization of QTLs. Construction of consensus genetic map and QTL meta-analysis were performed using BioMercator v3.1 (Sosnowski et al. 2012). a Rice chromosome 8 showing QTLs for grains per panicle (GPP), percent seed set (PSS), and osmotic adjustment (OA) across rice genetic backgrounds. The QTLs qGPP8.2 (IR64/Cabacu) and qPSS8.2 (IR64/Cabacu) were from Fig. 1 of this study, OA from the mapping population Co39/Moroberekan (Lilley et al. 1996), OA from IR62266/IR60080 (Robin et al. 2003), and OA from CT9993/IR62266 (Nguyen et al. 2004). b Rice chromosome 1 showing the common QTLs for grain yield (GY), leaf rolling (LR), osmotic adjustment (OA) and plant height (PH) under drought stress across rice genetic backgrounds. The QTLs shown are (A) GY from IR64/Cabacu from Fig. 1 of this study, (B) GY from N22/MTU1010 (Vikram et al. 2011), (C) GY from IR64/Azucena (Lafitte et al. 2002), (D) GY from CT9993/IR62266 (Kumar et al. 2007), (E) GY from Zhenshan97/IRAT109 (Yue et al. 2005), and (F) GY from CT9993/IR62266 (Babu et al. 2003); (G) LR from IR64/Cabacu from Fig. 1 of this study; (H) LR from Bala/Azucena (Price et al. 2002), and (I) LR from IR64/Azucena (Courtois et al. 2000); (J) OA from Co39/Moroberekan (Lilley et al. 1996) and (K) OA from IR62266/IR60080 (Robin et al. 2003); and (L) PH from IR64/Cabacu from Fig. 1 of this study
