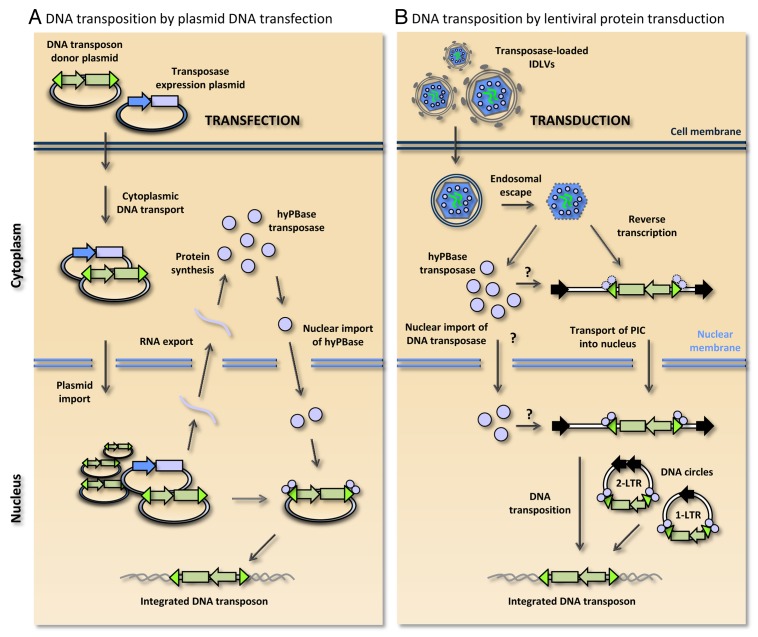
Figure 1. Schematic comparison of piggyBac DNA transposition by plasmid DNA transfection and lentiviral protein transduction. (A) DNA transposition by co-transfection of the DNA transposon donor plasmid and transposase-encoding plasmid. Transport through the cytoplasm and nuclear uptake lead to production of hyPBase transposase, which is subsequently imported into the nucleus. Within the nucleus the transposon-based gene vector (indicated in green) is excised from the donor plasmid and inserted into a genomic locus. (B) DNA transposition by lentiviral protein transduction in integrase-defective lentiviral vectors (IDLVs). Engineered lentiviral particles carry both the hyPBase protein (indicated by small light-purple circles) and the diploid RNA vector genome (indicated by green lines). Cell entry mediated by the VSV-G surface protein occurs through endocytosis and subsequent endosomal escape. Reverse-transcribed double-stranded DNA intermediates serve as transposon donors. Along with linear DNA substrates, 1-LTR and 2-LTR circles generated by homologous recombination and non-homologous end joining, respectively, may serve as transposon donors. Question marks indicate that it is not currently known whether transposase subunits are associated with the transposon in the cytoplasm or are imported into the nucleus prior to association with the transposon terminal repeats. It is currently unclear whether the transposase remains part of the pre-integration complex (PIC) during nuclear entry or is released from the PIC during cytoplasmic transport.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.