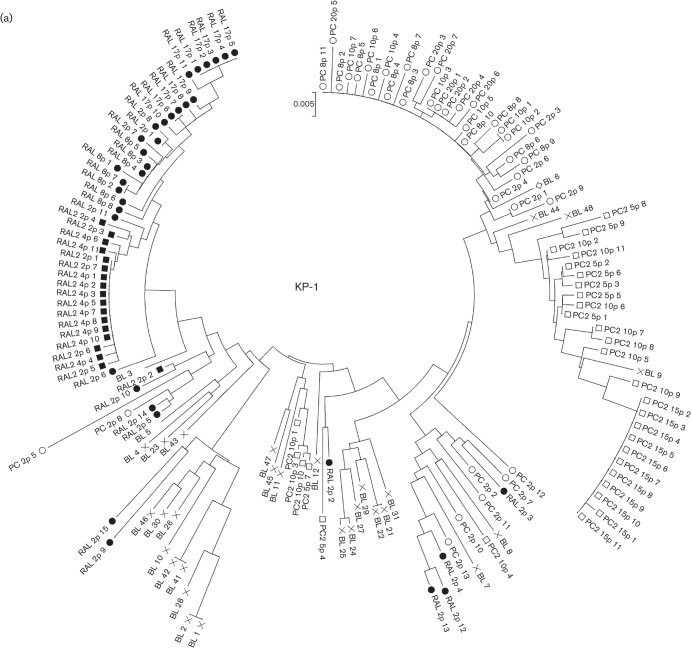
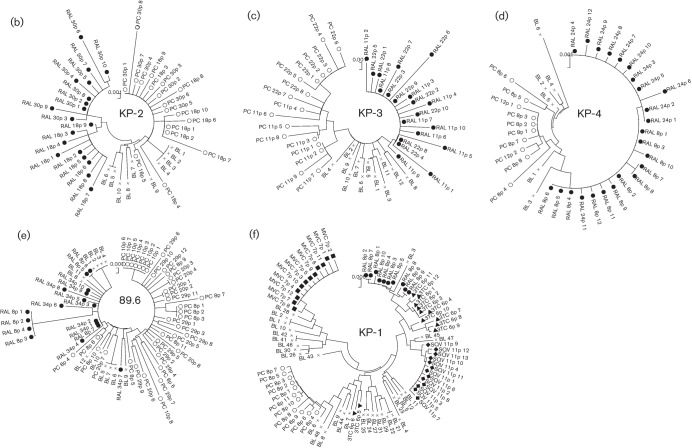
Fig. 2.
Phylogenetic analyses of the Env regions from in vitro-passaged viruses selected with or without ARV drugs. (a–e) Phylogenetic trees were constructed using gp120 SP–V5 sequences from RAL-selected and passage-control variants of KP-1 (a), KP-2 (b), KP-3 (c), KP-4 (d) and strain 89.6 (e). An ‘×’ represents baseline (BL) variants, and closed and open symbols represent RAL-selected (RAL) and passage-control (PC) variants, respectively. In (a), the results of the second experiment are indicated as RAL2 and PC2, respectively. (f) A phylogenetic tree was constructed using gp120 SP–V5 sequences from RAL-, 3TC-, SQV-, MVC-selected and control-passaged variants of KP-1. ○, Control variants after eight passages; •, RAL-selected variants after eight passages; ▴, 3TC-selected variants after six passages; ⧫, SQV-selected variants after 11 passages; ▪, MVC-selected variants after seven passages. The trees were constructed using the neighbour-joining algorithm embedded within the mega software.