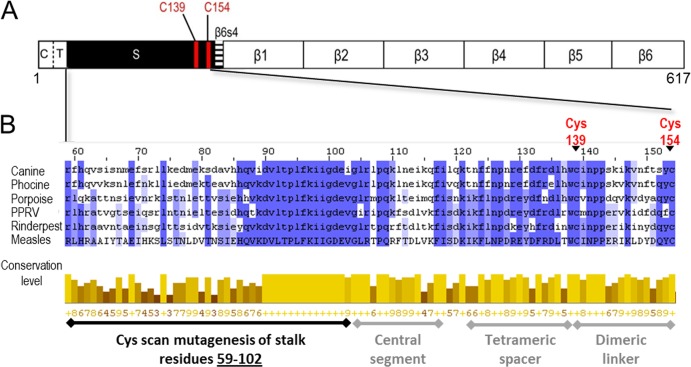
FIG 1.
Schematic of MeV H protein and sequence alignment of MeV H stalk to other morbilliviruses. (A) H linear structure. From left to right: C, cytoplasmic tail; T, transmembrane region; S, stalk; and β1 to 6, beta sheets 1 through 6. The two Cys residues that cross-link the H dimer are shown as red lines. (B) Sequence alignment of the MeV H stalk (residues 59 to 154) to five other morbilliviruses. The alignment was made using ClustalW2 (42). Canine, canine distemper virus; Phocine, phocine distemper virus; Porpoise, porpoise morbillivirus; PPRV, peste-des-petits-ruminants virus; Rinderpest, rinderpest virus; Measles, measles virus. (Top) Shades of blue represent the degree to which the identity and nature of an amino acid is conserved at a given position. (Bottom) The conservation level of each residue is indicated by the height of the bars below each residue and the associated score below the bar (0 to 9). The black horizontal line indicates the Cys mutants reported in this study. The gray horizontal lines indicate stalk segments, the structure and function of which were defined in a previous study (24).