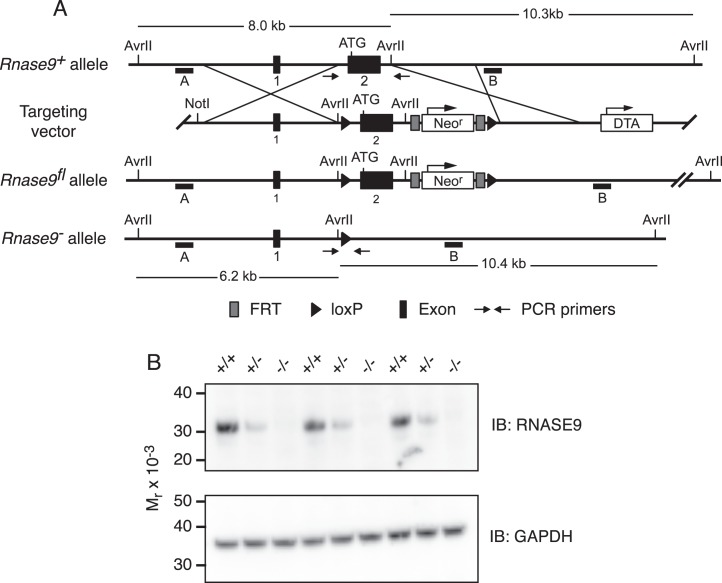
FIG. 1.
Rnase9 gene targeting scheme and confirmation of gene targeted deletion. A) Diagram of the wild-type Rnase9+ allele (top), targeting vector (top middle), Rnase9fl floxed allele (bottom middle), and Rnase9− null allele (bottom). The numbered black boxes indicate exons and the initiating ATG is shown near the 5′ end of exon 2. The 4.7-kb 5′ homology arm and the 2.9-kb 3′ homology arm are indicated. The positive selection neomycin resistance gene (Neor) and the diphtheria toxin A (DTA) negative selection cassette are shown with arrows indicating direction of transcription. Black triangles are loxP recombination sites, and dark gray boxes flanking the Neo cassette are flippase recognition target (FRT) recombination sites. The small horizontal black bars labeled A and B indicate the 5′ and 3′ external probes used for Southern blot analysis, respectively. AvrII restriction sites and predicted AvrII fragment sizes are shown. Small arrows indicate the locations of PCR primers used to detect the Rnase9+ and Rnase9− alleles. B) RNASE9 protein expression. Detergent extracts of mouse epididymis were prepared and analyzed by Western blot analysis using RNASE9 antisera as described in Materials and Methods.