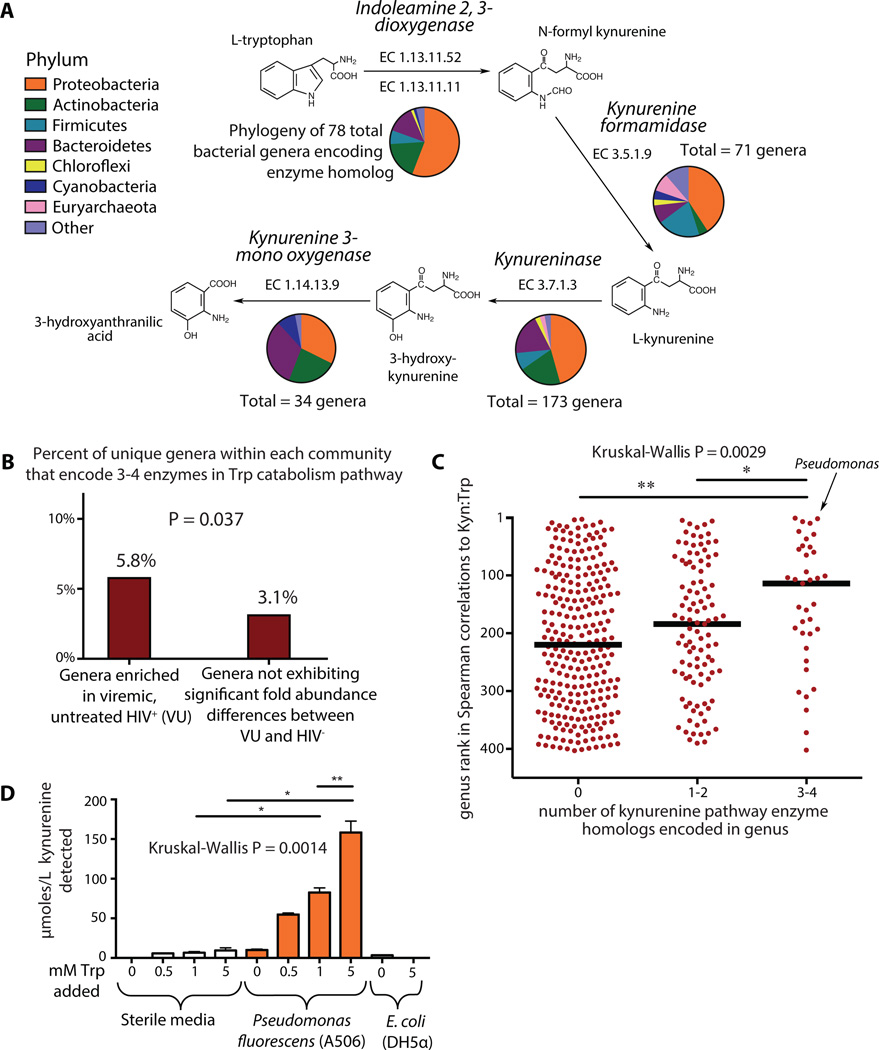
Figure 5. Bacterial tryptophan catabolism machinery is genetically and functionally homologous to IDO1 enzymatic activity and is enriched in the DMC.
(A) Homologs to tryptophan catabolism pathway enzymes are genetically encoded in numerous bacterial genera as denoted by ‘Totals.’ Phylogenetic distribution of annotated enzymes in the kynurenine pathway was collated and represented using pie charts as defined in legend. (B) A Fisher’s exact test for enrichment found that a greater proportion of genera overrepresented in VU encoded 3–4 genes involved in the tryptophan catabolism pathway as compared to genera not differing abundance between VU and HIV- (P = 0.037). (C) All genera that were both detected in our cohort and contained members for which genome sequence data was available were ranked based on their strength of non-parametric Spearman correlation to Kyn:Trp ratios in HIV-infected subject plasma. Genera with 3 or 4 genetically encoded homologs to kynurenine pathway enzymes exhibited significantly stronger correlations to plasma Kyn:Trp ratios than those with only 1 or 2 homologs, and even more so than those with no annotated homologous enzymes. (D) Pseudomonas fluorescens strain A506 and a lab strain of E. coli (DH5α) were grown in King’s B Medium for 20 hours in the presence of varying concentrations of tryptophan. Supernatants from these cultures were assayed for the presence of kynurenine. Error bars indicate standard deviations calculated from three independent experiments. Non-parametric Mann-Whitney tests were used for group comparisons (*P < 0.05, **P < 0.005).