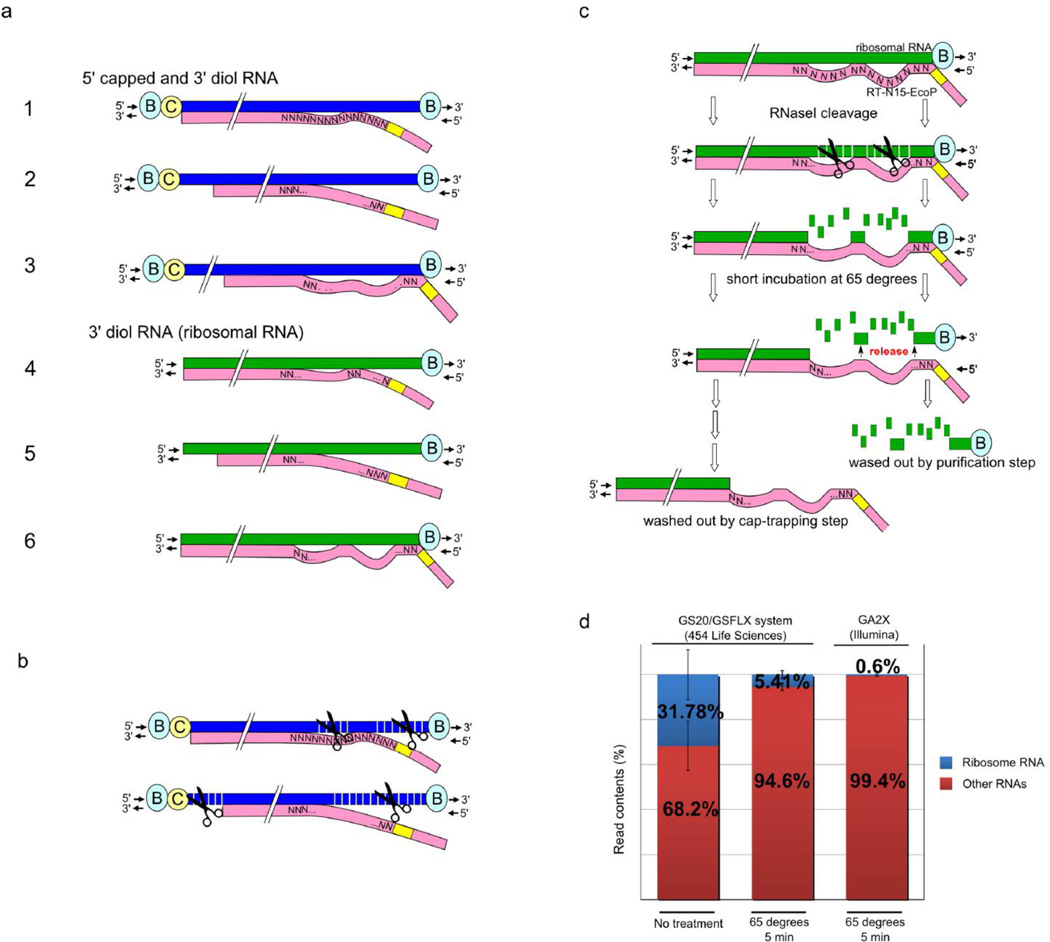
Figure 2. Strategies to eliminate non-capped biotinylated molecules.
(a) As well as at the 5’ end of capped RNAs, biotinylation takes place also on the diol group at the 3’ end of capped RNAs and at the 3’ end of ribosomal/other uncapped RNAs, which must be subsequently eliminated to avoid contamination of 5’ complete cDNA. Careful usage of random primers has been instrumental in achieving this. Blue strand indicates 5’ capped RNA and green strand indicates non capped RNA. Pink strand includes random primer (with restriction enzyme site in yellow) and shows first strand cDNA extension. Examples 1–6 show different potential random priming patterns. (C) Cap; (B) biotin. (b) RNase I is used to cleave single strand mismatched regions produced by cDNA synthesis using random primers. The two examples show different random priming patterns on 5’ capped RN; the upper example (from example 1 in panel a) results in capture of 5’ complete cDNA, while the bottom example (from example 2 in panel a) shows incomplete cDNA that did not extend to the 5’ end. The incomplete cDNA is subsequently eliminated due to RNase I cleavage from the biotinylated cap. (c) Uncapped/incomplete cDNAs derived from primers that perfectly matched the 3’ end of the RNA and biotinylated at the 3’ end need to be eliminated from the library to reduce bias due to ribosomal RNA contamination. Infrequent cases of perfectly aligned random priming at the 3’ end would cause capture through the 3’ end biotin on ribosomal RNA. However, long random primers (N15; pink) leave mismatches that are cleaved by RNAse I treatment, as described in (b). Heating to 65 °C after RNAse treatment releases the biotin at the 3’ end from the cDNA/RNA hybrid, which is then washed out at captrapping step. (d) Dramatic removal of ribosomal cDNA sequence tags by RNAse treatment and heating at 65°C. In a previous protocol using the GS20/GSFLX sequencer (454 Life Sciences)40, ribosomal CAGE tags represented ~30 % of the tags without treatment at 65°C (n = 6). Other samples were incubated 65 °C for 5 min resulting in ribosomal RNA decrease to 5.41 % (n = 6). Error bars indicate a standard deviation of experiments. A third sample shows further decreased ribosomal RNAs with the protocol here presented for the Illumina sequencing instruments. Other RNAs include RNA sequences mappable to the genome (60–89%) or unmapped RNA-derived sequences.