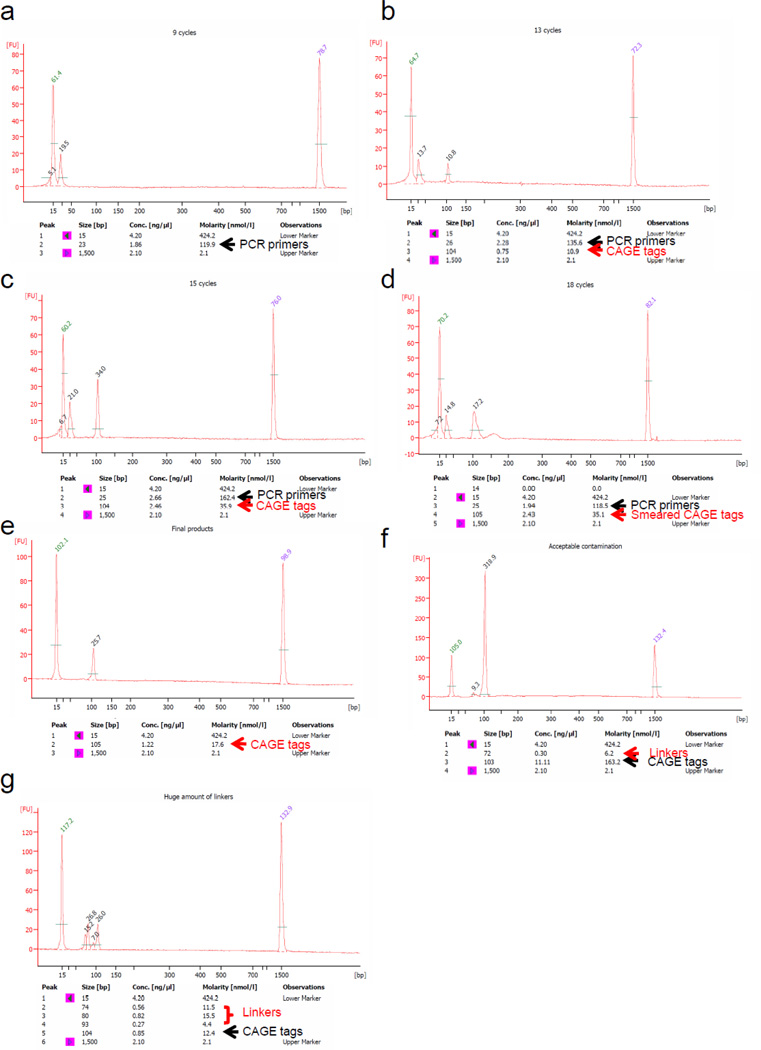
Figure 5. Measurement of PCR products.
Example of PCR cycle optimization by Agilent Bioanalyzer DNA 1000 kit. The amount of applied PCR product is 1 µl. (a) 9 cycles (b) 13 cycles, (c) 15 cycles, (d) 18 cycles. Peak values indicate the height of Fluorescence Units (FU). With only 9 cycles, there is only primer peak (25 bp) and the CAGE peak is not visible. With 13 cycles, there are two peaks, the primer peak and the CAGE peak. The measured size may slightly differ from the actual 96 bp within the inherent instrument error range (103 – 105 bp). CAGE tag peaks with FU values between 5 and 10 (molarity: ~10 nmol l−1) are suitable for bulk PCR. In case of 15 cycles, the FU exceeds 20 (molarity: ~30 nmol l−1) and with 18 cycles the reactions shows a broad peak, due to over cycling (compared to a and b). (e) Final product molarity of the single peak was estimated to be 17.6 nmol l−1 at 13 cycles. PCR primers are subsequently removed during Steps 55 and 57. After Step 58, the single peak products are ready for sequencing. (f) Example of small linker dimer contamination (70 – 80 bp), which does not affect sequencing, and large linker contamination (g), which prevents the usage of the library.