Figure 5. MSI-1 regulates stearoyl-CoA desaturase.
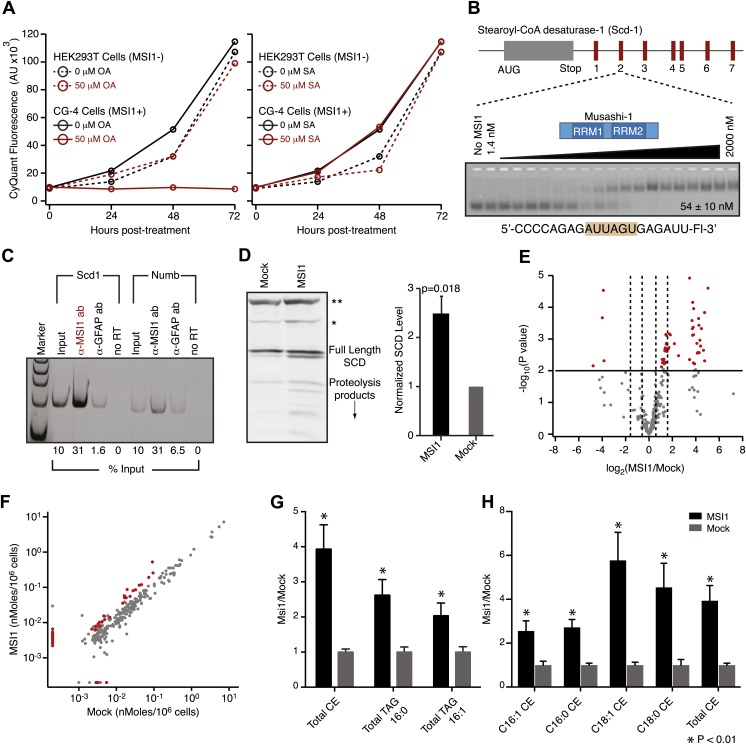
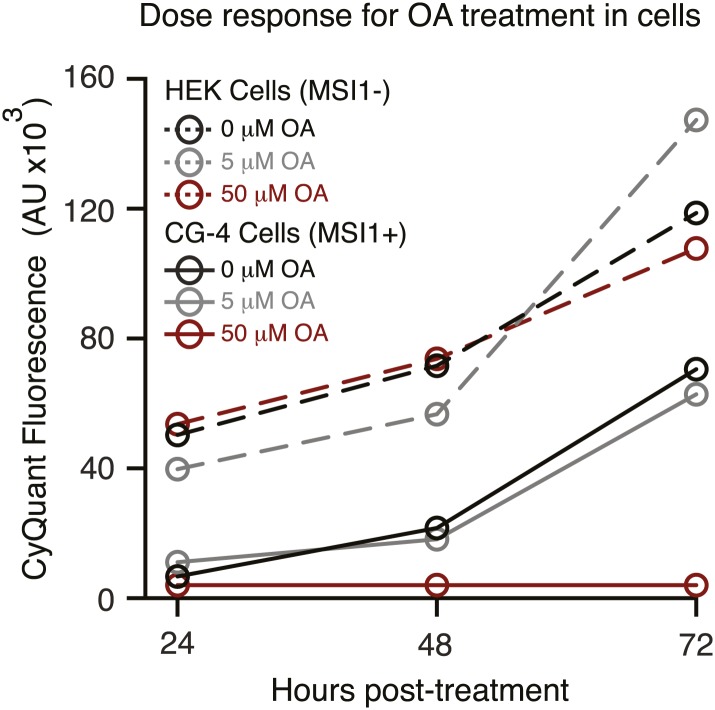
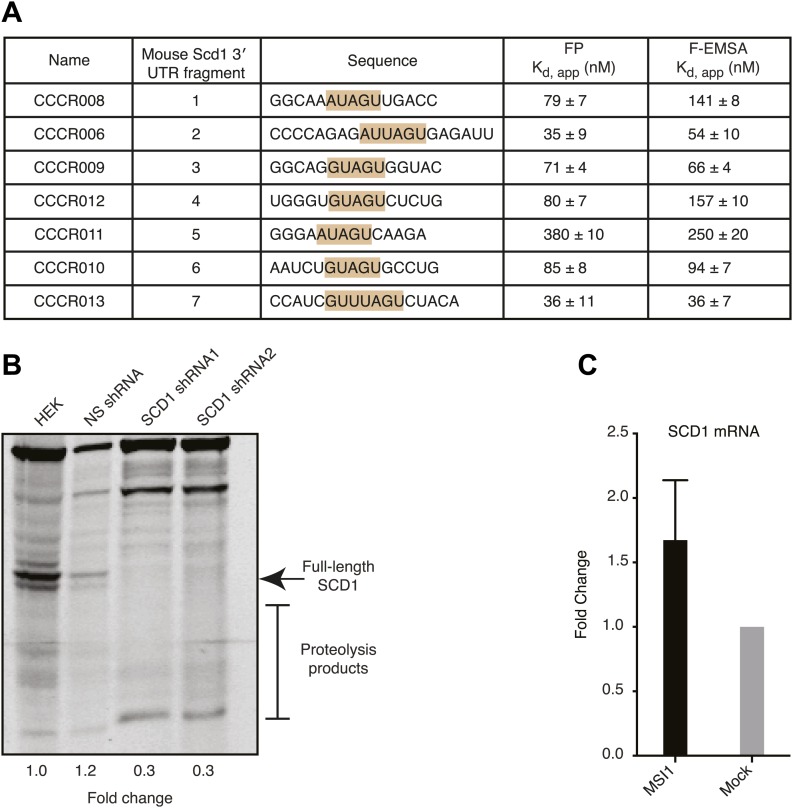
(A) HEK293T (dashed) and CG-4 (solid) cell proliferation as a function of oleic acid or stearic acid treatment (red = treated, black = untreated). The data are the average and standard deviation of at least three biological replicates. (B) There are seven MSI1 consensus sites in the 3′-UTR of Scd-1 mRNA. The Kd, app is the average and standard deviation of at least three experiments. (C) Scd-1 transcripts co-immunoprecipitate with anti-MSI1 antibodies. The data were quantified using a FUJI FLA-5000 imager. (D) Western analysis of SCD expression in HEK293T cells. The data were quantified using the LICOR Odyssey system relative to non specific bands (** and *, Figure 5—figure supplement 2B) to control for loading. The average and standard deviation of at least three independent experiments is shown. (E–H) Lipidomics analysis of HEK293T cells ± MSI1 expression. Source data are included in Figure 5—source data 1. (E) Volcano plot of lipidomics data. Dashed lines denote fold-changes of ±1.5 and ±3. Red data points indicate lipids that are significantly changed upon MSI1 expression. (F) Scatter plot of lipidomics data. Data are reported as nMoles per million cells. Red data points indicate lipids that are significantly changed upon MSI1 expression (FDR = 0.05). (G) Fold-changes of the total cholesterol esters and two TAG classes in which 38 of the 54 significantly changing lipids are categorized. Each class changes significantly with MSI1 overexpression (p<0.05). (H) Fold-changes for the four lipids that comprise the total cholesterol esters class. All display significant changes with MSI1 expression (FDR = 0.05).
DOI: http://dx.doi.org/10.7554/eLife.02848.014