Table 1.
Structure-activity relationship analysis demonstrates specificity of inhibition
| A | ||||
|---|---|---|---|---|
| Compound name | CID | Screen score | FP Ki, app (µM) | F–EMSA Ki, app (µM) |
| Chembridge 7409829 | 28425 | 0.045 | 15 ± 2.8 | 54 ± 22 |
| Aurintricarboxylic Acid (ATA) | 2259 | 0.053 | 0.23 ± 0.03 | 1.5 ± 0.14 |
| GW7647 | 3397731 | −0.028 | 6.5 ± 0.4 | 21 ± 0.8 |
| Oleic Acid | 445639 | −0.005 | 1.2 ± 0.4 | 1.4 ± 0.7 |
| B | ||||
|---|---|---|---|---|
| Compound name | Structure | Code | FP Ki, app (µM) | F–EMSA Ki, app (µM) |
| Oleic acid | 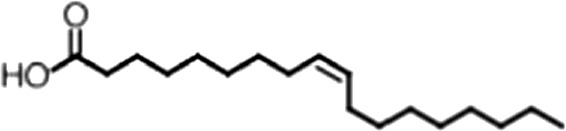 |
18:1 ω-9 | 1.2 ± 0.4 | 1.4 ± 0.7 |
| Eicosenoic acid | 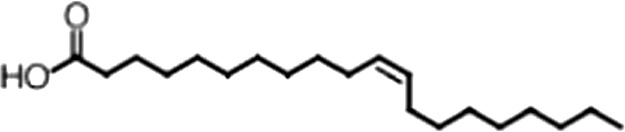 |
20:1 ω-9 | 1.2 ± 0.4 | 1.7 ± 0.6 |
| Erucic acid | 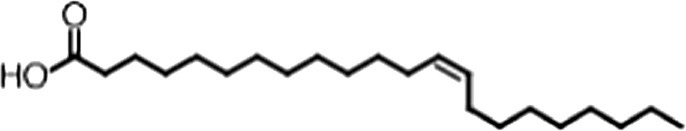 |
22:1 ω-9 | 0.64 ± 0.2 | 0.82 ± 0.03 |
| Nervonic acid | 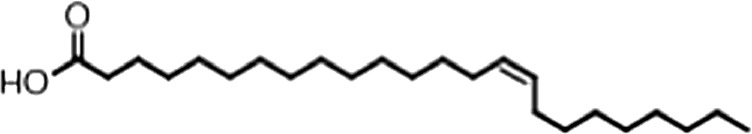 |
24:1 ω-9 | 47 ± 30 | 23 ± 8 |
| Palmitoleic acid | 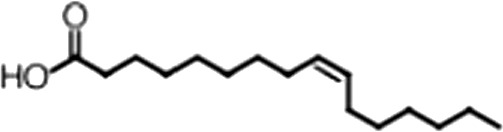 |
16:1 ω-7 | 5.3 ± 0.5 | 13 ± 0.9 |
| Linoleic acid | 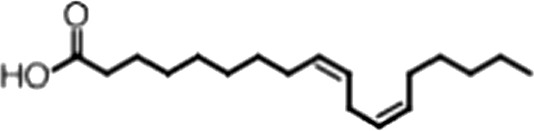 |
18:2 ω-6, 9 | 2.2 ± 0.2 | 1.2 ± 0.03 |
| Arachidonic acid | 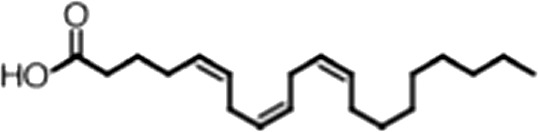 |
20:4 ω-6, 9, 12, 15 | 3.0 ± 0.2 | 1.1 ± 0.3 |
| Oleoyl-CoA | 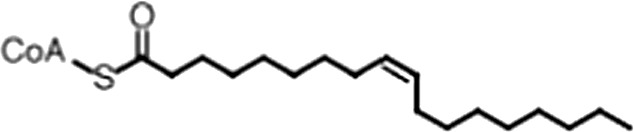 |
(18:1 ω-9) | 8.1 ± 0.3 | 4.0 ± 0.2 |
| Erucyl-CoA |  |
(18:1 ω-9) | 4.1 ± 0.9 | 0.62 ± 0.2 |
| Ricinoleic acid | 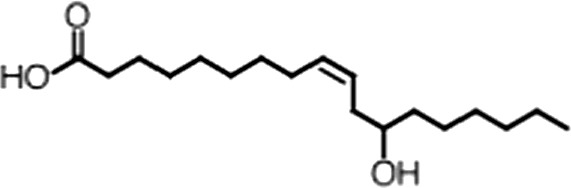 |
(18:1 ω-9) | No inh. | 18 ± 9 |
| Oleamide | 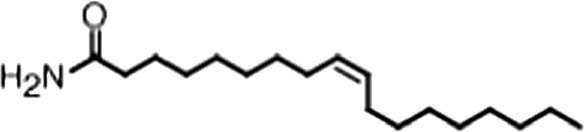 |
(18:1 ω-9) | No inh. | No inh. |
| Ethyl oleate | 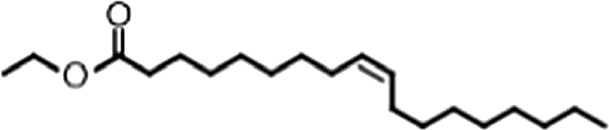 |
(18:1 ω-9) | No inh. | No inh. |
| 4-Methylumbelliferyl Oleate | 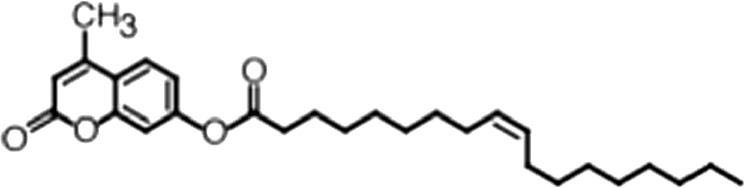 |
(18:1 ω-9) | No inh. | No inh. |
| Elaidic acid | 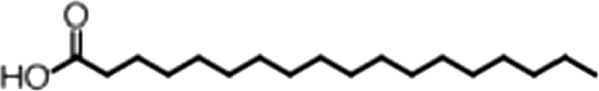 |
18:1(trans) | No inh. | No inh. |
| Stearic acid | 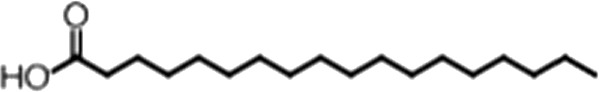 |
18:0 | No inh. | No inh. |
| Palmitic acid | 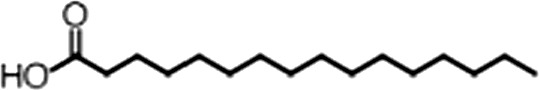 |
16:0 | No inh. | No inh. |
| Myristic acid |  |
14:0 | No inh. | No inh. |
(A) Small molecule screen hits. Compound ID (CID) refers to each compound's LOPAC identification number. Screen scores were calculated by normalizing the polarization value of each compound to the no protein and no compound controls, as described in the supplemental methods. After the screen was complete, compounds that scored as hits were confirmed by FP and F–EMSA dose response experiments. Apparent inhibition constants (Ki, app) are the average and standard deviation of at least three independent experiments. (B) The code = carbon number:number of double bonds, followed by the position of the double bonds from the aliphatic end of the fatty acid. Where a fatty acid is modified, the parental fatty acid numerical code is given in parentheses for comparison purposes. FP and F-EMSA dose response results are reported as the average and standard deviation of at least three independent experiments.
