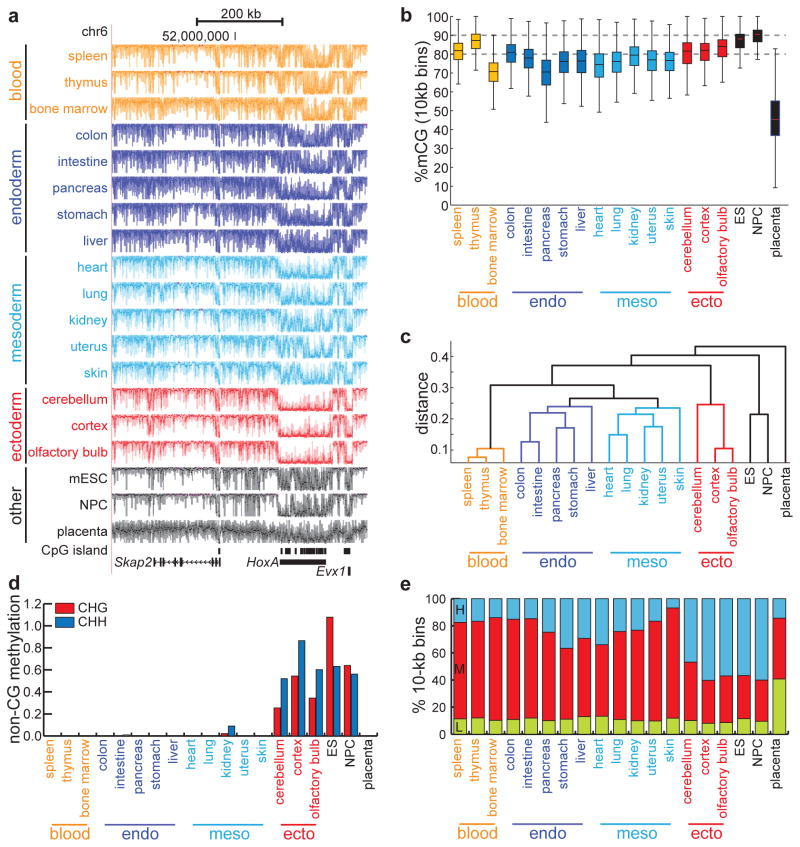
Figure 1. Distinct tissue-specific methylomes.
(a) UCSC Genome Browser snapshot of DNA methylomes, derived from tissues of a single mouse, near the HoxA locus. Each track spans %mCG values between 0% and 100%. Tracks are colored by tissue type, as determined by the clustering in (c). (b) Boxplots of the %mCG distributions for each tissue, calculated from non-overlapping 10-kb bins spanning the mouse genome. Boxplot edges indicate the 25th and 75th percentiles, and whiskers indicate non-outlier extremes. (c) Dendrogram constructed from 1-kb regions exhibiting significant tissue-specific methylation (p = 0.001, χ2 test). Distance is measured as 1 − Pearson correlation coefficient. (d) Global abundance of methylation in non-CG context, expressed as the difference between the fraction of non-CG cytosines that are methylated and the bisulfite non-conversion rate. (e) Abundance of large domains bearing low (L), medium (M), or high (H) methylation, as determined by hidden Markov models independently trained on each tissue.