Abstract
Immunologic dysregulation is the cause of many non-infectious human diseases such as autoimmunity, allergy and cancer. The gastrointestinal tract is the primary site of interaction between the host immune system and microorganisms, both symbiotic and pathogenic. Here we discuss findings which indicate that developmental aspects of the adaptive immune system are influenced by intestinal bacterial colonization. We also highlight the molecular pathways that mediate host–symbiont interactions that regulate proper immune function. Finally, we present recent evidence to support an emerging concept whereby disturbances in the bacterial microbiota result in immunological dysregulation that may underlie disorders such as inflammatory bowel disease. Perhaps the mammalian immune system which appears designed to control microbes is, in fact, controlled by the microbes themselves.
Humans represent a scaffold upon which diverse microbial ecosystems are established. Immediately after birth, all mammals are initiated into a life-long process of colonization by foreign microorganisms that inhabit most environmentally exposed surfaces (such as the skin, mouth, gut and vagina)1,2. Shaped by millennia of evolution, some host–bacterial associations have developed into beneficial relationships creating an environment for mutualism. A key example of such an environment is provided by the vast numbers and diversity of bacteria that are harboured by the lower gastrointestinal tract of mammals3,4,1,5. By young adulthood, both humans and other mammals support one of the most complex microbial ecosystems on the planet, with over 100 trillion bacteria comprised of potentially many thousands of microbial species in the distal gut6,7. Symbiotic bacteria of the mammalian gut have long been appreciated for the benefits they provide to the host: through provision of essential nutrients, metabolism of indigestible compounds, defence against colonization by opportunistic pathogens and even contribution to the development of the intestinal architecture8. Moreover, it seems that certain basic developmental features and functions of the mammalian immune system depend on interactions with the human microbiome9. Unlike opportunistic pathogens, which elicit immune responses that result in tissue damage during infection, some symbiotic bacterial species have been shown to prevent inflammatory disease during colonization (see below). Surprisingly, the ‘normal’ microbiota also contains microorganisms that have been shown to induce inflammation under particular conditions. Therefore, the microbiota has the potential to exert both pro- and anti-inflammatory responses, and balances in the community structure of gut bacteria may be intimately linked to the proper function of the immune system.
By virtue of its responsibility to recognize, respond and adapt to countless foreign and self molecules, the immune system is central to the processes of health and disease. While the classical view of the immune system is that it evolved to protect organisms from infection by microbial pathogens, we peacefully co-exist with our vast and complex microbiota. In fact, the magnitude of the molecular interactions between the microbiota and the host immune system appear to be extensive. Here, we discuss recent evidence that suggests a beneficial partnership has evolved between symbiotic bacteria and the immune system. These molecular interactions seem to direct the development of immune responses, and in turn, the immune system shapes the composition of the microbiota. We highlight seminal examples of microorganisms that have a role in preventing inflammatory bowel diseases (IBD), and the beneficial immune responses they elicit during protection. Furthermore, technological advances now allow a more detailed understanding of the alterations of the microbial population of the gut during IBD. If some bacteria are actively shaping a healthy immune system, does the absence of these organisms lead to disease? It has recently been proposed that the mammalian genome does not encode the information (i.e., molecules) capable of carrying out all functions required for health, and that products of our microbiome (the collective genomes of the microbiota) are crucial for protection from various diseases10. It is possible that alterations in the development or composition of the microbiota (known as dysbiosis) disturb the partnership between the microbiota and the human immune system, ultimately leading to altered immune responses that may underlie various inflammatory disorders in humans.
Insights gained from germ-free mice
Developmental defects in germ-free mice
Several important effects of the microbiota on the host immune system have been determined by studies of gnotobiology, which is the selective colonization of germ-free (sterile) animals. Immune responses in germ-free animals can be considered naïve to the ‘education’ provided by molecules of both pathogenic and beneficial microorganisms. Germ-free animals show extensive deficits in the development of the gut-associated lymphoid tissues (GALT)9,11. In addition to numerous defects in antibody production (Box 1), germ-free animals have relatively fewer and smaller Peyer's patches and mesenteric lymph nodes (MLNs) compared to animals housed under specific pathogen free (SPF) conditions (Table 1). A recent report has shown that germ-free animals display impaired development and maturation of isolated lymphoid follicles (ILFs)12. These inducible structures seem to form normally following the introduction of gut bacteria, suggesting a dynamic relationship between the immune system and the microbiota. Together with various morphological tissue defects observed in the intestines of germ-free animals, it appears that the entire ultrastructural development of the gut is intimately connected to intestinal bacteria. For example, intestinal epithelial cells (IECs), which line the gut and form a physical barrier between luminal contents (including the microbiota) and the underlying cells of the immune system, have altered patterns of microvilli formation and decreased rates of cell turnover in germ-free animals compared with wild-type animals13. Gut bacteria have been shown to direct the glycosylation of lumenally-exposed surface proteins of the epithelium14. IECs have many immunological functions (Table 1): they can secrete and respond to various cytokines and express molecules (such as major histocompatibility molecules) that directly interact with lymphocytes. Expression and localization of pattern-recognition receptors (e.g., Toll-like receptors; TLRs) by the epithelium is influenced by bacterial colonization of the gut, and expression of defensins and other antimicrobial proteins are deficient in germ-free animals (Table 1). Consistent with this notion, the Gram-negative commensal organism Bacteroides thetaiotaomicron, but not the Gram-positive microbe, Bifidobacterium longum, induces the expression of the antimicrobial peptide, RegIIIγ, by specialized IECs known as Paneth cells15, 16. Intriguingly, the specificity of RegIIIγ is directed toward specific Gram-positive bacteria. It is therefore tempting to speculate that symbiotic bacteria direct innate immune responses of the gut in an effort to protect their environment. Collectively, these observations of developmental defects in germ-free mice at the tissue, cellular and molecular levels suggest that ‘normal’ immune function may be impaired in the absence of the microbiota.
Table 1.
Intestinal immunologic defects in germ-free mice
| Intestinal organ development | Site | Phenotype in Germfree mice |
|---|---|---|
| Small Intestine | Peyers Patches | fewer, less cellular |
| Lamina propria | thinner, less cellular | |
| Germinal centers | fewer plasma cells | |
| Isolated lymphoid follicles | smaller, less cellular | |
| Mesenteric Lymph nodes | Germinal centers | smaller, less cellular |
| fewer plasma cells |
| Cellular Defects | Cell Type | Phenotype in Germfree mice |
|---|---|---|
| Intestinal epithelial lymphocytes | CD8+ T cells | fewer, reduced cytotoxicity |
| Lamina propria lymphocytes | CD4+ T cells | proportional decrease in number |
| decreased Th17 cells (Small intestine) | ||
| increased Th17 cells (Colon) | ||
| Mesenteric lymph nodes | CD4+CD25+ T cells | reduced expression of Foxp3 |
| reduced suppressive capacity |
| Molecular immune deficiencies | Molecule | Phenotype in Germfree mice |
|---|---|---|
| Paneth Cells | Angiogenin-4 | reduced expression |
| RegIIIγ | reduced expression | |
| B cells | Secretory IgA | reduced production |
| Intestine | ATP | reduced |
| Intestinal epithelial cells | MHC class II | reduced expression |
| TLR 9 | reduced expression | |
| IL-25 | elevated |
Deficiencies in immune responses in germ-free mice
Germ-free animals are more susceptible to infection by certain bacterial, viral and parasitic pathogens. When challenged with the Gram-negative enteric pathogen Shigella flexneri, germ-free animals showed decreased immune resistance to infection and increased mortality compared with conventionally colonized animals17. Prior colonization with specific commensal bacteria antagonized S. flexneri infections, whereas colonization with control species such as E.coli did not, implying that some members of the microbiota provide protection against intestinal bacterial pathogens18. Infection by the Gram-positive intracellular pathogen Listeria monocytogenes results in decreased bacterial clearance in germ-free compared with colonized animals19.The mechanism for this increased susceptibility has been attributed to a T cell trafficking defect in germ-free animals to the site of Listeria infection. L-selectin and CD44 are known to be involved in homing of lymphocytes to sites of inflammation. Listeria infected germ-free mice have decreased accumulation of CD44+L-selectin+ T cells resulting in increased bacterial burden compared to SPF animals20. Salmonella typhimurium is known to cause a more severe acute gastroenteritis in germ-free animals21, however the reasons for this remain unclear. Establishing an infection requires the initial task of colonizing the host. For intestinal pathogens this can pose a difficult problem as all mammals are stably colonized by a consortium of bacteria that can act as a barrier to infection (“colonization resistance”). Recent studies suggest that inflammation induced in response to S. typhimurium changes the composition of the microbiota and suppresses its regrowth. Moreover, S. typhimurium exploits this deficiency in “colonization resistance” to establish infection and cause disease22. While important for maintaining a barrier to the colonization of potentially pathogenic organisms, it appears that the microbiota may also provide the host immune benefits. Supporting this, germ-free animals show reduced antigen-specific systemic immune responses to S. typhimuirum23. These studies support the idea that enteric pathogens such as S. typhimurium may have developed strategies to counter both the immune system and the microbiota during the infectious process.
Although significant work is still required to determine the beneficial immune responses induced by the microbiota, it is exciting to consider the teleological notion that indigenous bacteria actively prevent enteric disease by infectious microorganisms to fortify their niche. If true, then an evolutionary alliance has been forged between mammals and beneficial bacteria that is critical in maintaining the long-term survival of both. In other words, is our wellbeing dependent on the microbes we harbour?
The microbiota and inflammatory bowel disease
Immune system regulation during IBD
The impact of the microbiota on human health is best exemplified by studies in IBD, such as Crohn's disease and ulcerative colitis24-26. Both represent serious medical disorders marked by aberrant inflammation within the human gastrointestinal (GI) tract, resulting in severe clinical outcomes in affected patients. The causes for these diseases are complex, and include the contributions of genetic, geographic and habitual factors27. IBD (particularly Crohn's disease) is generally thought to be driven by T lymphocytes. The disease has classically been characterized by increases in pro-inflammatory cytokines such as tumour-necrosis factor (TNF) and interferon-γ (IFNγ). Recently, a new population of inflammatory T cells, termed T helper 17 (TH17) cells, have been implicated in the pathogenesis of human and experimental colitis28-33These cells are characterized by their expression of the pro-inflammatory cytokine interleukin-17 (IL-17), and require IL-23 for their maintenance and function. Specialized T cells, known as regulatory T (TReg) cells act to counter-balance these pro-inflammatory responses. FOXP3 is a transcription factor believed to be the ‘master regulator’ of TReg cells, and its absence results in massive multi-organ lymphoproliferative disease34. The mechanisms by which Tregs are able to suppress inflammation are quite diverse. These include the expression of inhibitory cytokines such as IL-10, TGF-β, and IL-35; disruption of cellular metabolism by expression of the IL-2 receptor, CD25; cytolysis, and targeting the maturation of DCs through surface expression of molecules such as CTLA-4 and LAG-335. A population of intestinal dendritic cells expressing the surface antigen CD103 have recently been demonstrated to be instrumental in the development and function of intestinal Foxp3+ Tregs. CD103+ DCs, but not CD103− DCs are able to ‘convert’ CD4+Foxp3− T cells into CD4+Foxp3+ Tregs cells in a TGF-β and retinoic acid dependent manner36, demonstrating that specialized mechanisms exist within the intestine to promote induction and maintenance of Tregs.
The importance of Tregs in regulation of intestinal homeostasis is best demonstrated by the ability of these cells to prevent induction of experimental colitis upon transfer into diseased hosts 37. The ability of Tregs to secrete IL-10 and IL-35 has been reported to be important during protection. Indeed, Treg cells deficient in either of the two subunits of IL-35 (Ebi3 and IL-12a), are unable to cure experimental colitis38. Additionally, animals in which the anti-inflammatory cytokine IL-10 has been specifically ablated from CD4+FOXP3+ T cells succumb to inflammatory disease of the intestine (as well as skin and lung), but show no signs of autoimmunity. Thus, it appears that cytokine production by Treg cells may be a primary protective mechanism that limits uncontrolled immune responses at environmentally-exposed surfaces such as the gut.
Recent evidence has begun to reveal mechanisms of intestinal immune modulation by the microbiota. Germ-free animals are deficient in the development of TH17 cells in the small intestine39. In this report, the reduction in IL-17 production is reciprocally coupled to an increase in CD4+FOXP3+ TReg cells in the germfree colon. Reconstitution of animals with a complex and diverse microbiota that does not contain the prominent phyla Bacteroidetes is unable to restore proper immune balance, suggesting that discrete organisms may have the capacity to modulate pro- and anti-inflammatory responses in the gut. The identity of specific bacterial species and particular molecules with dedicated functions to regulate T-helper/TReg-cell profiles in the gut remain unknown. However, ‘general’ bacterial products appear to have immunomodulatory affects. ATP produced by intestinal bacteria specifically increases colonic IL-17 production (and not IFNγ production)40. Consistent, with this, germ-free animals have reduced IL-17 and ATP levels in the colon. Additionally, commensal bacterial DNA triggers TLR9 to confer resistance to the enteric parasite Encephalitozoon cuniculi 41. Antibiotic treatment of animals to eliminate gut bacteria results in increased susceptibility to infection by this parasite, consistent with findings from studies in germ-free mice. Treatment of infected animals with DNA extracted from the intestinal microbiota upregulated Th1 and Th17 responses, while suppressing Treg activity resulting in decreased parasite burden. Previous work had implicated TLR signaling as being important for gut homeostasis42; therefore these recent findings extend this observation to suggest that a molecular dialogue between immune receptors and microbial molecules confers resistance to enteric infection.
The contribution of the microbiota to the development of TRegcells remains unclear as multiple studies have revealed conflicting results. An earlier report demonstrated a deficiency in the percentage of Foxp3+ cells within the CD4+CD25+ subset within the MLNs of germfree mice when compared to conventionally colonized animals. Foxp3 mRNA levels were also lower in CD4+CD25+ isolated from total lymph nodes of germfree mice. Supporting this, another study reported lower expression of the Foxp3 transcript in CD4+CD62L− T cells from germfree mice.43. Additionally, studies have shown that TReg cells from germ-free animals were not as potent in suppressing CD4 T-cell proliferation in vitro as cells from conventionally colonized animals44. Populations of TReg cells from germ-free animals expressed less IL-10, and were unable to prevent disease in a transfer model of experimental colitis45. In contrast, recent studies have reported no change in the percentage of CD4+Foxp3+ subset of T cells within the colonic lamina propria46, while yet another study reported elevated percentages of CD4+TCRβ+Foxp3+ within the small intestine39. These differences might be attributable to the specific subsets of TReg analyzed, differences in experimental methodologies and/or the tissues from which TReg were harvested. Alternatively, the particular diet given to the animal might influence Treg subsets within the intestine as most animal food, even if autoclaved, may have varying amounts of microbial molecules (such as TLR ligands). However, these data collectively suggest that intestinal bacteria interact with the mammalian immune system to direct the linage differentiation of both pro- and anti- inflammatory T-cell populations. Therefore, induction of effector T-cell responses and modulation of TReg -cell function by the microbiota may be a crucial component of diseases such as IBD. It is possible that different classes (or even species) of bacteria induce diverse immunological functions. Therefore, the equilibrium between inflammation and regulation in the gut may be due to the community structure of the microbiota.
IBD and a breakdown in tolerance to gut bacteria
IBD involves a shift from a regulated intestinal immune responses to one that is driven by unrestrained immune-cell activation and pro-inflammatory cytokine production47,48,49. The cause of this increase in immune stimulation is of great interest, and several lines of evidence indicate a significant role for commensal bacteria in the progression of disease 50. Patients with IBD respond favorably to antibiotic treatment and fecal diversion, and have greater antibody titers against indigenous bacteria than unaffected individuals51,52,53. In addition, inflammatory lesions are more pronounced in areas of the intestine that contain the greatest number of bacteria. The data in animal models provide further evidence for the involvement of gut bacteria in IBD. Pre-treatment with antibiotics has been shown to alleviate intestinal inflammation in several animal models54,55. HLA-B27-transgenic rats, interleukin-10 (IL-10)-and IL-2-deficient mice spontaneously develop chronic colitis, whereas germ-free animals of all backgrounds fail to develop intestinal inflammation56,57,58. In a model of disease induced by the adoptive transfer of pathogenic T cells into immunodeficient (Scid−/− or Rag−/−) recipient mice, colonization of animals with intestinal pathogens such as Helicobacter hepaticus was found to exacerbate inflammation59. Moreover, pathogenic T cells can be transmitted to healthy animals through the adoptive transfer of T cells that are reactive against specific commensal organisms50,60. The only organism reported to be strongly associated with Crohn's disease is adherent-invasive E.coli (AIEC)61. However, it appears that inflammatory responses during human and experimental IBD are directed towards certain subsets of commensal organisms with pathogenic potential such as Helicobacter, Clostridium and Enterococcus species. Curiously, these organisms are abundant commensals, and not typically infectious pathogenic agents. As the microbiota of all mammals contains these potentially harmful species, known as pathobionts (commensals with pathogenic potential), the reason why inflammation ensues only in subjects affected by IBD is not entirely known. Genetic factors play an important role in the pathogenesis of IBD. This is demonstrated by familial aggregation of IBD and increased concordance for IBD in monozygotic twins. Genome wide association studies have identified genetic defects which are highly linked to disease. Mutations in bacterial sensing (NOD2/Card15)62 and T cell immunity (IL-23R)30 have highlighted the connection between microbes and inflammation in IBD. Additionally, experimental systems have demonstrated the importance of host genetics during induction of IBD. Indeed, microorganisms such as E.coli and E. faecalis, that do not initiate disease in an immunologically competent host, do so when introduced into genetically pre-disposed strains 63. Some have predicted that IBD, at least in part, results from an imbalance in the normal microbiota (termed dysbiosis) without acquisition of an infectious agent64. It remains unclear whether dysbiosis directly causes disease or is a result of the altered intestinal environment. Future studies using animal models whereby the microbiota can be selectively manipulated during the course of experimental disease may begin to address this important issue.
Genetic and habitual/lifestyle factors play a crucial role in maintaining health, however, the notion that dysbiosis influences intestinal disease has gained much attention5,65,66. Recent studies have now begun to show evidence that the composition of the microbiota alone (and not genetics or environment) may be important to the induction of disease. T-bet (tbx21) is a T-box transcription factor that controls the type 1 pro-inflammatory response in both adaptive and innate immune cells67. Loss of T-bet in mice lacking an adaptive immune system (Rag−/−) develop spontaneous and penetrate intestinal inflammation that resembles ulcerative colitis68. Treatment of these mice with broad spectrum antibiotics cured the intestinal disease, demonstrating that inflammation was driven by the microbiota. Finally, when wild-type animals are co-housed with T-bet−/−Rag−/− colitic mice, they develop a comparable colitis like disease68. Therefore, the transfer of colitigenic microbes alone (the identity of which is still uncertain) was sufficient to induce experimental ulcerative colitis. The second study addressing the sufficiency of dysbiosis have come from metagenomic analysis of the microbiota during obesity. Ob/ob mice display symptoms of obesity and are used as a model system of human disease. Remarkably, transfer of the microbiota from ob/ob animals into germ-free wild-type animals results in an increase in the mean body fat of recipient animals69,70. Moreover, the proportion of Bacteroidetes is decreased in obese people when compared to lean people, demonstrating the possibility that alterations in the human microbiota may affect disease. Much like IBD, obesity appears to have a strong genetic component71. The complex interplay between host genotype and its effects on the microbiota are an area very worthy of further investigation. However, studies such as these, where bacterial transfer into wild-type animals induces , argue for a prominent role for dysbiosis.
Similar to animal studies, dysbiosis has been implicated in human disease. Many investigations have shown a significant alteration in the microbiota of patients with IBD 72,73,74. A recent metagenomic (culture-independent analysis of microbial community structure) case-control study compared the microbiota of patients with IBD to that of non-IBD controls74. The results revealed a statistically significant difference between the microbial compositions of IBD patients and non-IBD patient controls. Furthermore, the authors reported that an ‘IBD-specific’ microbiota displays a reduction in the levels of two phyla of bacteria, the Firmicutes and Bacteriodetes, both prominently represented in non-IBD controls. Longitudinal studies are required to determine if this particular profile (that is, the loss of certain classes of bacteria) can be used as a diagnostic tool to identify people with a greater likelihood of developing IBD. Although our understanding of how dysbiosis may affect IBD is still preliminary, novel sequencing technologies promise to provide a platform to analyze many thousands of microbiomes of both healthy and diseased individuals75. With increased knowledge of species-specific alterations during disease, the molecular mechanisms that link dysbiosis of the microbiota to intestinal inflammation can systematically be explored in both experimental and human studies.
Beneficial gut bacteria promote immune homeostasis
The evidence presented so far has implicated the microbiota in shaping immune system responses during disease, but the question still remains as to which particular organisms are mediating these beneficial responses and, more importantly, how this is achieved. Here we review the mechanisms by which symbiotic bacteria affect the mammalian immune system that result in the prevention or treatment of experimental and human IBD.
In the early 1900s, Ilya Mechnikov was the first to propose the use of live microorganisms to maintain bowel health and prolong life. Now, the term ‘probiotic’ is used to describe dietary microbes that are able to confer a health benefit on the host76. As shown in Table 2, many individual or combinations of bacterial species have been shown to ameliorate the symptoms of IBD in humans and mouse models. Although many of these probiotic strains decrease toxic microbial metabolic activities, more recent evidence demonstrates the capacity of these organisms to modulate intestinal immune responses. The common feature of almost all bacterial species used as probiotics is the ability of these organisms to control inflammation. Bacterial species can act on several cell types (epithelial cells, dendritic cells (DCs) and T cells), but recent evidence suggests that induction of regulatory T cells by these organisms is crucial to limiting inflammation and disease. Treatment of colitic mice with the probiotic cocktail, VSL#3, increased the production of IL-10 and the percentage of TGFβ-expressing T cells77. More importantly, transfer of lamina propria mononuclear cells from VSL#3-treated mice was able to prevent colitis in recipient mice, indicating the ability of the VSL#3 cocktail to initiate the generation of a protective population of cells. Depletion of TGFβ-bearing CD4+ T cells from probiotic treated mice before the transfer of lamina propria cells abolished the protective capacity of these cells77. More recently, in a model of pathogen-induced inflammation, treatment of mice with Bifidobacteria infantis led to a downregulation of intestinal inflammation and increases in the number of CD4+CD25+ TReg cells78. Adoptive transfer of the CD4+CD25+ TReg-cell population from mice fed with B. infantis inhibited inflammation-induced activation of nuclear factor-κB (NF-κB) in recipient mice.
Table 2.
Bacteria shown to be protective in inflammatory bowel disease
| Bacterial Strain/s | Model system | IBD type/ model | Mechanism of suppression |
|---|---|---|---|
| VSL#3 | Human/ mouse | Pouchitis, Ulcerative colitis/ TNBS | IL-10, TGF-β bearing T cells |
| Bifidobacteria lactis | Rat | TNBS | ↓ colonic TNF-α and iNOS |
| Bifidobacteria infantis | Mouse | S. typhimurium induced enteritidis | Tregs, inhibits NF-κB |
| E. coli Nissle 1917 | Human/ mouse | Ulcerative colitis/ DSS colitis | ↓ inflammation via TLR-2 & TLR-4 |
| Lactobacillus rhamnosus GG | Mouse/ Rat | TNBS/HLA-B27 | induction of CD4+CD25+ cells |
| Lactobacillus salivarius | Mouse | TNBS | ↓ colonic inflammation |
| Lactobacillus reuteri | Mouse | IL-10−/− mice | NGF and ↓ IL-8, TNF in cell lines |
| Lactobacillus plantarum 299v | Mouse | IL10 −/− mice, but not TNBS | ↓ IFN-γ and IL-12p40 |
| Lactobacillus fermentum | Rat | TNBS | ↓ colonic TNF-α and iNOS |
| Lactobacillus casei | Rat | TNBS | ↓ colonic COX-2 |
| Bacteriodes thetaiotaomicron | Rat | S.enterica induced enteritidis | ↓ IL-8 and TNF-α in Caco-2 cells |
| Bacteriodes fragilis | Mouse | T cell transfer model, TNBS | CD4+ T cell derived IL-10 |
| YO-MIX Y109 FRO 1000 | Mouse | TNBS | ND |
| Faecalibacterium prausnitzii | Mouse | TNBS | ↓ NF-κB and IL-8 |
Naive CD4+ T cells can adopt a regulatory phenotype through interactions with intestinal DCs36. Foligne et al.79 demonstrate that bone-marrow-derived DCs (BMDCs) internalize Lactobacillus rhamnosus, yet maintain an immature phenotype. Transfer of BMDCs incubated with L.rhamnosus can protect from inflammation and disease when transferred into a recipient animal with colitis79. Moreover, depletion of the CD4+CD25+ T-cell subset abolished the ability of the probiotic-treated DCs to protect from disease, suggesting that L. rhamnosus-treated DCs can initiate TReg-cell activity79. It has recently been shown that some patients with Crohn's disease showed a specific reduction in a prominent gut microbe, Faecalibacterium prausnitzii80. Intriguingly, this organism or its secreted substances were able to induce anti-inflammatory responses (such as IL-10 expression) and ameliorate induction of TNF-α and intestinal disease when orally administered to experimental animals. This seminal study thus directly linked the numerical reduction of a beneficial bacterium from the human microbiota to the development of disease, suggesting that symbiotic microbes may be directly mediating health. The specific molecules made by these particular bacterial species that guide immune responses remain unknown; however current data support the idea that symbiotic organisms actively communicate with the host immune system to modulate anti-inflammatory processes.
The first demonstration that a single molecule made by a commensal microorganism could shape beneficial immune responses was provided by the identification of polysaccharide A (PSA) produced by the human symbiont Bacteroides fragilis (Figure 1). Colonization of germ-free mice with B. fragilis or treatment with purified PSA directs the cellular and physical development of the immune system, including expansion and differentiation of splenic CD4+ T cells81. The novel immunomodulatory activities of PSA during B. fragilis colonization of germ-free mice include correcting systemic T-cell deficiencies, T-helper cell imbalances and directing lymphoid organogenesis. The importance of B. fragilis in maintenance of a healthy immune response was recently illustrated by the finding that colonization by B. fragilis or treatment with purified PSA can protect from the induction of experimental IBD66. Moreover, mice that are colonized by a mutant form of this microorganism that lacks expression of PSA (B. fragilis ΔPSA) are no longer protected from disease. Oral treatment of mice with purified PSA protects recipients from weight loss, decreases levels of the pro-inflammatory cytokines TNFα, IL-17 and IL-23, and inhibits epithelial hyperplasia and neutrophil infiltration to the gut associated with disease induction in these models66. To provide insight into the mechanistic basis for PSA-mediated protection, it was shown that increases in local induction of IL-10 were required for the anti-inflammatory properties of PSA. PSA is unable to protect from induction of colitis in IL-10-deficient mice, demonstrating that PSA functions by inducing the production of IL-10.. Based on this observation, we tested the ability of PSA to induce IL-10 in CD4+ T cells. Indeed, CD4+ T cells purified from MLNs during PSA-mediated protection of colitis showed increased levels of IL-10 compared with PBS treated control mice. Finally, transfer of IL-10-deficient CD4+ T cells into Rag−/− recipient mice abolished the ability of PSA to protect from experimental colitis66. These data have identified a single molecule made by a symbiotic microorganism that stimulates CD4+ T cells to make IL-10, required for the suppression of the inflammatory process during colitis, and suggest that other beneficial bacteria may produce factors that can positively shape the host immune response during IBD.
Figure 1. Model for Bacteroides fragilis-mediated protection from disease induced by Helicobacter hepaticus.
B. fragilis produces an immunomodulatory polysaccharide (PSA) that induces an immunoregulatory programme that provides protection from inflammation induced by H. hepaticus. PSA is taken up by intestinal dendritic cells (DCs), which presumably migrate to the local mesenteric lymph nodes (MLNs) where they initiate T-cell responses by presenting PSA on MHC class II molecules to CD4+ T cells. This process helps to restore a balanced T helper (TH) and regulatory T (TReg) cell profile. Subsequently, naive TH cells adopt anti-inflammatory functions that include expression of interleukin-10 (IL-10). IL-10 is required to suppress the production of pro-inflammatory cytokines (such as IL-17, IL-23 and tumour-necrosis factor (TNF) induced by H. hepaticus during experimental colitis. It is this balance of the pro-inflammatory responses to H. hepaticus by regulation induced by B. fragilis that results in the control of intestinal inflammation.
For many years, IL-10-producing CD4+ T cells (so called Tr1 cells) were considered to be distinct from ‘natural’, thymic-derived TReg cells that are characterized by the expression of FOXP382. It is now apparent that there is overlap between these two populations and that IL-10-producing regulatory T cells can be found in the Foxp3+ T cell subset and are imperative for the control of experimental colitis. While IL-10 producing T cell clones specific for cecal bacterial contents have been generated 83the ability of a molecule from symbiotic bacteria to regulate FOXP3+ TReg-cell differentiation and function awaits further validation. However, it appears that certain beneficial bacteria have evolved molecules (known as symbiosis factors) that induce protective intestinal immune responses. The current treatments for IBD are either ineffective in most patients or result in severe side effects. Knowledge of which beneficial species of bacteria can prevent or cure disease, and harnessing the potent immunosuppressive potential of symbiosis factors will be important steps towards designing novel and natural therapeutics for IBD.
The microbiota hypothesis and human disease
Does harboring certain strains of bacteria predispose an individual to disease or protect from it? B. fragilis has been shown to protect its host from inflammatory disease caused by H. hepaticus in an animal model of experimental colitis66. As symbiotic bacteria seem to have evolved mechanisms to promote protection from pathobionts found in the same microbiota, does disease result from the absence of specific organisms and their beneficial molecules? In other words, if symbiosis factors actively maintain health, can dysbiosis reduce bacterial-mediated immune regulation that subsequently leads to inflammation? Reflecting a growing medical crisis in Western societies, recent epidemiological and clinical reports have revealed dramatic increases in the incidences of several immune disorders. Incidences of IBD, asthma, atopic disorders (skin, respiratory, and food allergies), rheumatoid arthritis, type 1 diabetes and multiple sclerosis are increasing dramatically in ‘westernized’ populations, including those of western European nations, the United States and Japan. The rapidity of the rise in disease rates appears to argue against a solely genetic basis for these observations 84. Because the implementation of antimicrobial strategies (including vaccination, sanitation, ‘western’ diets, and antibacterial therapeutics) does not permit discrimination between infectious and non-infectious microorganisms, such interventions have most likely led to changes in our association with the microbial world as a whole.
If improvements in hygiene and health care have altered the process by which a healthy microbiota is assembled and maintained, then patients with these diseases in developed countries should display signs of dysbiosis. This indeed appears to be the case, at least according to a growing number of studies which are now linking ‘Western’ diseases to alterations in the microbiota. The intestinal bacterial composition of IBD patients differs greatly from that of healthy controls74. However, over many years, research to identify the pathogenic organism(s) that elicit inflammation has repeatedly identified reactions only to intestinal bacteria shared by all humans—healthy and ill. No infectious organisms have ever been conclusively demonstrated to be involved in IBD, though various studies are still ongoing. So, perhaps the targets of inflammation in IBD are not pathogens, but pathobionts that are overrepresented during dysbiosis (Figure 2). When intestinal bacterial species were analyzed as a factor in asthma development, reports concluded that allergic children from both an industrialized nation (Sweden) and a developing country (Estonia) had lower levels of colonization by Bacteroides spp. and higher levels of colonization by aerobic microorganisms than did non-allergic children from either region85. Epidemiological studies have linked an altered intestinal microbiota to other allergic disorders, such as atopic eczema and rheumatoid arthritis86-88. Though cause or effect relationships still remain unresolved, it seems that deviations in the composition of the gut microbiota may be an influencing environmental factor underlying the development of disease in genetically predisposed individuals.
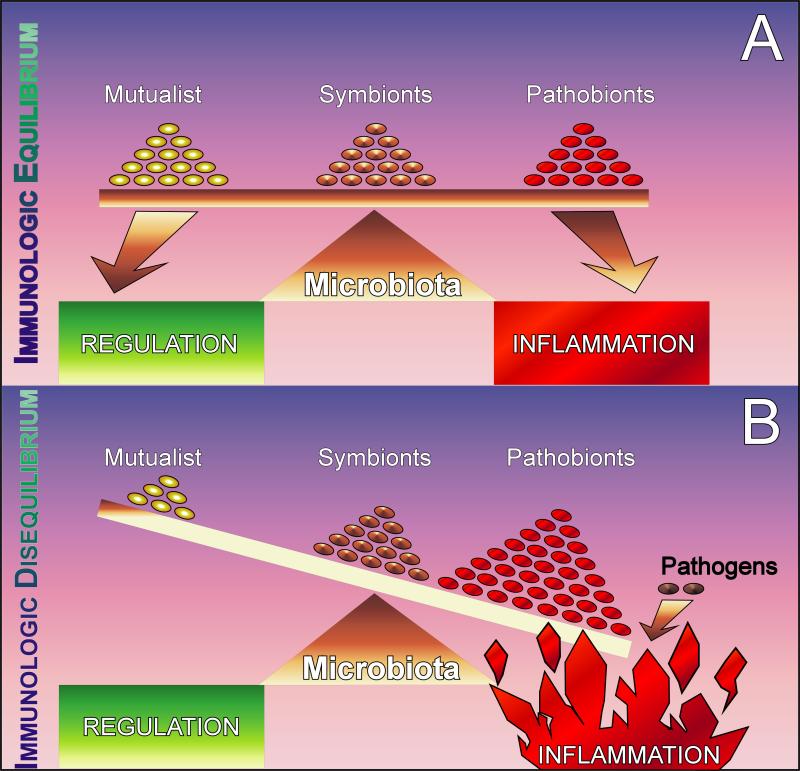
Figure 2. Immunological dysregulation is the result of dysbiosis in the microbiota.
a | A healthy microbiota contains a balanced composition of multiple classes of bacteria. Commensals are permanent residents of this complex ecosystem and provide no benefit or detriment to the host (at least to our knowledge). Symbionts are organisms with known health-promoting functions. Pathobionts are also permanent residents of the microbiota with the potential to induce pathology. b | During dysbiosis, there is an unnatural shift in the composition of the microbiota whereby either the numbers of symbionts are reduced and/or pathobionts are increased. The various causes for this are not entirely clear, but are likely to include recent societal advances in developed countries. The result is non-specific inflammation which may predispose certain genetically susceptible people to inflammatory disease. Pathogens are opportunistic organisms that cause rare and acute inflammation.
Recent studies have launched a revolution in biology aimed at understanding how (and, more importantly, why) mammals harbour multitudes of symbiotic bacteria. As discussed above, the effects of the microbiota on the immune system are becoming increasingly evident. Astonishingly, the immune disorders for which incidences have increased in ‘Western’ countries all seem to involve reduced TReg-cell activity. It has been shown in animal models and some human studies that deficiencies in TReg-cell populations or function underlie asthma, IBD, rheumatoid arthritis, type 1 diabetes and multiple sclerosis89. CD4+CD25+FOXP3+ TReg cells can prevent, and in some cases, treat these disorders in laboratory animals. The antagonism between pro-inflammatory TH17 cells and FOXP3+ TReg cells is well documented90. Most remarkably, the numbers and function of certain TReg-cell populations are reduced in germ-free animals45,43. As mentioned above, germ-free animals have numerous other immunological defects that may lead to disease, which implicates a role for the microbiota in actively supporting health. After millions of years of co-evolution with our microbial partners, have societal advances paradoxically affected human health adversely by reducing our exposure to health-promoting bacteria?
Implications for a ‘healthy’ immune system
Although it has been known for decades that we harbour millions of commensal bacteria, recent studies have only just begun to reveal the extraordinary complexity and diversity of the human microbiota. This consortium of bacteria contains tenfold more cells than the human body, 100 times the number of genes than in the human genome and has the metabolic capacity of the human liver91,92. How is such a complex microbial network assembled after birth? Relman, Brown and co-workers recently examined the development of the intestinal microbiota of infants93. They found that in the first few days to weeks of life, the microbiota of newborns is highly variable and subject to waves of temporal fluctuations, possibly representing a time of sampling, or ‘trial and error’, to coordinately assemble a stable microbiota. The first years of life are also a time of significant post-natal development of the immune system. As commensal gut bacteria have profound influences on the immune system, deviations from the ‘normal’ development of the microbiota (through modern strategies such a caesarian section, formula-based diet, hygiene, vaccination, and use of antimicrobials in infants) may alter the outcome of immune development and potentially predispose individuals to various inflammatory diseases later in life (Figure 2).
Based on clinical, epidemiological and immunological evidence, it seems possible that changes in the intestinal microbiota may be an essential factor in the incidence of numerous inflammatory disorders. Intestinal bacteria are a critical component in instructing the development and function of the immune system. It is conceivable that the absence of beneficial microorganisms (due to dysbiosis) that promote appropriate immune development leads to the inflammatory responses that underlie various immune diseases in humans. Recent studies have shown that at least for experimental IBD, spontaneous disease results when immune suppression is deficient; thus inflammation appears to be a default immunological state in the absence of regulation94. Although pathogenic bacteria clearly induce local inflammation during acute infections, have symbiotic bacteria evolved to regulate those inflammatory processes that are harmful to the host (and therefore, harmful to the existence of the symbiont)? Significant research has implicated innate and adaptive immune suppression during the control of disorders such as IBD, autoimmunity, asthma and allergy, cancer and infectious diseases. According to a wealth of recent studies, we propose the notion of a vast, intricate and unexpected level of interdependence between beneficial bacteria and the immune system. Perhaps genetic and habitual factors shape the composition of the microbiota, which in turn shapes the immune system of individuals predisposed to inflammatory disease (Figure 3). The emerging identification of human symbiotic bacteria with potent anti-inflammatory properties, and their correlative absence during disease, suggests that certain aspects of human health may depend on the ‘health’ of the microbiota. The medical and social reconsideration of the microbial world may have profound consequences for the health of our future generations.
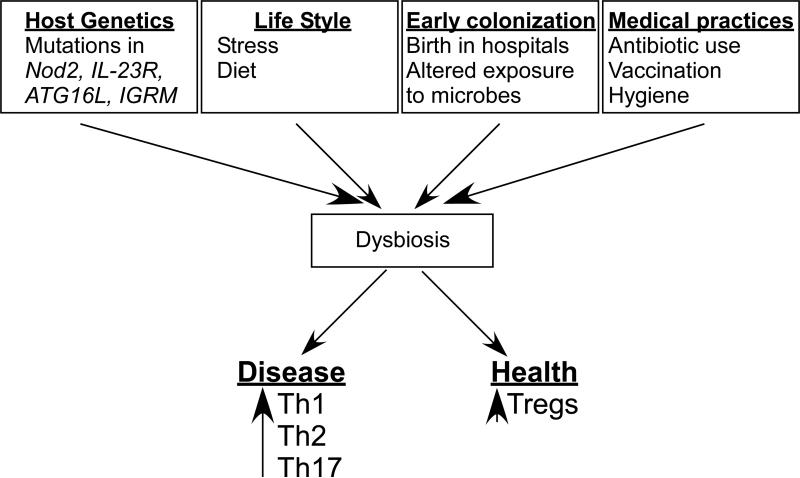
Figure 3. Proposed causes of Dysbiosis.
We propose the notion that the composition of the microbiota can shape a healthy immune response or predispose to disease. Multiple factors can contribute to a dysbiotic state including host genetics, medical practices, life style, and exposure. a| Host genetics can potentially influence dysbiosis in multiple ways. Genome wide association studies of IBD patients have revealed polymorphisms in immune-related genes. An individual lacking genetic regulatory mechanisms or overactive pro-inflammatory pathways will have an intestinal environment marked by unchecked inflammation. Studies have demonstrated the ability of inflammation alone to influence the composition of the microbiota, potentially in favor of pathobionts. Alternatively, it is possible that genetics of the host can ‘select’ or exclude the colonization of particular organisms. This selection can be either active (as would be the case of an organism recognizing a particular receptor on the host) or passive (the host environment is more conducive to fostering the growth of select organisms). ‘Selection’ of pathobionts by the host could tip the balance in favor of inflammation. b| The food we consume and even the day-to-day stress95 represent aspects of life style that have the potential to influence the microbiota. c| Hospitals represent a sterile environment in which infants are born. While the sterility protects from dangerous pathogens, it also has the potential to prevent early exposure to health-promoting bacteria. d| The advent of antibiotics represents a major medical breakthrough; however, antibiotics do not have the capacity to distinguish between pathogenic or symbiotic microorganism s and may adversely alter the microbiota.
Antibody responses in germ-free animals.
One of the first immunologic deficiencies observed in germ-free mice was a profound reduction in the levels of secretory immunoglobulin A (sIgA) found in the intestine96. Mono-association of mice with various bacteria leads to increased IgA expression. As numerous studies have demonstrated that sIgA coats commensal (and pathogenic) bacteria, some have speculated IgA is involved in limiting the penetration of bacteria into host tissues. Studies from activation-induced cytosine deaminase (AID)-deficient animals (which are unable to class switch to IgA), display lymphoid hyperplasia of the gut and an altered microbiota, favoring the outgrowth of specific classes of bacteria97. Though experimental evidence does indicate that IgA is involved in protection from some enteric bacterial and viral infections98, contradictory studies have shown that IgA deficiency does not cause increased prevalence of disease in animals, and IgA-deficient people generally live a healthy life. However, pioneering discoveries initially by Macpherson and colleagues are revealing a mechanistic role for symbiotic bacteria in actively shaping the production of sIgA. Dendritic cells (DCs) that have acquired gut commensal bacteria migrate to MLNs where they induce the production of IgA from naïve B cells99. This process is required to control the penetration of commensal bacteria through the gut epithelial barrier100. Recently, it has been shown that sensing of symbiotic bacterial products by TLRs of intestinal epithelial cells results in localized B cell class switching101, a process that potentially affects immune interactions with the microbiota102. The discovery that symbiotic bacteria direct the function of a specialized mucosal DC population that induces IgA class switching also implicates the microbiota in having evolved defined mechanisms to direct immune responses103. Further, IgA responses were recently shown to be involved in maintaining host-bacterial mutualism by limiting innate immune responses to a model gut symbiont104. The recent revitalization of research into the biological functions of intestinal sIgA promises to provide important clues regarding the molecular communication between the immune system and the microbiota.
Acknowledgements
We thank members of the Mazmanian laboratory for their critical review of the manuscript. The authors apologize to those whose work could not be mentioned due to space limitation and the focus of the manuscript, in particular the vast clinical data in IBD. J.L.R is a Merck Fellow of the Jane Coffin Child's Memorial Fund. S.K.M. is a Searle Scholar. Work in the laboratory of the authors is supported by funding from the National Institutes of Health, Damon Runyon Cancer Research Foundation and the Crohn's & Colitis Foundation of America to S.K.M.
Glossary
- Microbiota
The amalgam of microorganisms that make up a complex and diverse community living within a given anatomical niche (usually an environmentally exposed surface of the body).
- Microbiome
The collective genomes of a microbiota.
- Symbiosis
Symbiosis means “living together” as originally defined, the relationship must be constant, intimate and between dissimilar species. Although often used to describe a beneficial relationship, symbiosis literally does not imply that either partner gains an advantage.
- Symbiont
An organism that lives in association with a host (usually for a lifetime) without obvious benefit or harm to either member.
- Mutualism
A symbiotic association in which both members benefit from the relationship.
- Commensal
From the Latin, com mensa, meaning to ‘share a table’. This term is frequently used to describe a microorganism which benefits from an association with no known effects on the host.
- Pathogen
From the Greek, pathos, meaning ‘suffering’. An opportunistic organism that rarely comes in contact with the host, but causes acute or chronic disease upon infection.
- Parasite
An opportunistic organism that maintains a prolonged, close association with the host which benefits the parasite at the expense of the host.
- Pathobiont
A symbiont that does not normally elicit an inflammatory response but under particular conditions (environmentally induced) has the potential to cause dysregulated inflammation leading to disease.
- Dysbiosis
Changes in the ‘normal’ or healthy composition of the microbiota due to lifestyle practices that alter the initial development or stable maintenance of the microbiota.
- Specific pathogen free (SPF)
Typically used as conventionally colonized animals, these animals are reared and maintained in an environment with an unknown complex microbiota, that is free from known pathogens.
- Gut-associated lymphoid tissues
Lymphoid structures and aggregates associated with the intestinal mucosa, specifically the tonsils, Peyer's patches, lymphoid follicles, appendix or cecal patch and mesenteric lymph nodes. They are enriched in conventional and unconventional lymphocytes and specialized dendritic-cell and macrophage subsets. They provide the first line of defence against entry of pathogens through the mucosal barrier.
- Peyer's patches
Groups of lymphoid nodules present in the small intestine (usually the ileum). They occur massed together on the intestinal wall, opposite the line of attachment of the mesentery. Peyer's patches consist of a dome area, B-cell follicles and interfollicular T-cell areas. High endothelial venules are present mainly in the interfollicular areas.
- Mesenteric lymph nodes
(MLNs). Lymph nodes located at the base of the mesentery. They collect lymph (including cells and antigens) draining from the intestinal mucosa.
- Cryptopatches
Clusters of cKIT+IL-7Rα+Thy1+ T-cell progenitors found in the crypt lamina propria of both small and large intestinal villi.
- Isolated lymphoid follicles
Small lymphoid aggregates located in the anti-mesenteric wall of the small intestine and containing B cells, dendritic cells, stromal cells and some T cells. They may contain germinal centres. They are thought to have a role in maintaining equilibrium between the immune system and the microbiota.
- Pattern-recognition receptor
(PRR). A host receptor (such as Toll-like receptors) that can sense pathogen-associated molecular patterns and initiate signaling cascades (which involve activation of nuclear factor-κB) that lead to an innate immune response.
- Inflammatory bowel disease
(IBD). A chronic condition of the intestine that is characterized by severe inflammation and mucosal destruction. The commonest forms in humans are ulcerative colitis and Crohn disease.
- Crohn's disease
A form of chronic inflammatory bowel disease that can affect the entire gastrointestinal tract, but is commonest in the colon and terminal ileum. It is characterized by transmural inflammation, strictures and granuloma formation, and is believed to result from an abnormal T-cell-mediated immune response to commensal bacteria.
- Ulcerative colitis
A chronic disease characterized by inflammation of the mucosa and sub-mucosa of the large intestine
- ‘Natural’ Regulatory T cell
(TReg cell). A specialized type of CD4+ T cell that can suppress the responses of other T cells. These cells provide a crucial mechanism for the maintenance of peripheral self-tolerance and are characterized by expression of CD25 (the α-chain of the interleukin-2 receptor) and the transcription factor forkhead box P3 (FOXP3).
- VSL#3
A mixture of bacteria consisting of four strains of Lactobacillus (L. casei, L. plantarum, L. acidophilus, and L. delbrueckii subsp. bulgaricus), and three strains of Bifidobacterium (B. longum, B. breve, and B.infantis), and one strain of Streptococcus salivarius subsp. thermophilus.
Biography
Sarkis K. Mazmanian: Sarkis K. Mazmanian is an assistant professor in the Division of Biology at the California Institute of Technology. He was formerly an assistant professor at Brigham and Women's Hospital and Harvard Medical School. He received his doctoral training at the University of California, Los Angeles studying the mechanism by which Gram-positive pathogens anchor surface protein adhesins during bacterial infection. His work now focuses on the study of beneficial bacterial associations with mammalian hosts and its implications to health.
June L. Round: June L. Round is a postdoctoral fellow in the laboratory of Sarkis K. Mazmanian. She received her Ph.D from University of California, Los Angeles where she studied scaffolding molecules involved in T cell biology . Her more recent interests are how symbiotic bacteria within the intestine shape mucosal T cell development and responses.
References
- 1.Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006;124:837–48. doi: 10.1016/j.cell.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 2.Dethlefsen L, McFall-Ngai M, Relman DA. An ecological and evolutionary perspective on human-microbe mutualism and disease. Nature. 2007;449:811–8. doi: 10.1038/nature06245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hooper LV. Bacterial contributions to mammalian gut development. Trends Microbiol. 2004;12:129–34. doi: 10.1016/j.tim.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 4.Mazmanian SK, Kasper DL. The love-hate relationship between bacterial polysaccharides and the host immune system. Nat Rev Immunol. 2006;6:849–58. doi: 10.1038/nri1956. [DOI] [PubMed] [Google Scholar]
- 5.Peterson DA, Frank DN, Pace NR, Gordon JI. Metagenomic approaches for defining the pathogenesis of inflammatory bowel diseases. Cell Host Microbe. 2008;3:417–27. doi: 10.1016/j.chom.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frank DN, Pace NR. Gastrointestinal microbiology enters the metagenomics era. Curr Opin Gastroenterol. 2008;24:4–10. doi: 10.1097/MOG.0b013e3282f2b0e8. [DOI] [PubMed] [Google Scholar]
- 7.Ley RE, et al. Evolution of Mammals and Their Gut Microbes. Science. 2008 doi: 10.1126/science.1155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hooper LV, Gordon JI. Commensal host-bacterial relationships in the gut. Science. 2001;292:1115–8. doi: 10.1126/science.1058709. [DOI] [PubMed] [Google Scholar]
- 9.Macpherson AJ, Harris NL. Interactions between commensal intestinal bacteria and the immune system. Nat Rev Immunol. 2004;4:478–85. doi: 10.1038/nri1373. [DOI] [PubMed] [Google Scholar]
- 10.Zaneveld J, et al. Host-bacterial coevolution and the search for new drug targets. Curr Opin Chem Biol. 2008;12:109–14. doi: 10.1016/j.cbpa.2008.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Falk PG, Hooper LV, Midtvedt T, Gordon JI. Creating and maintaining the gastrointestinal ecosystem: what we know and need to know from gnotobiology. Microbiol Mol Biol Rev. 1998;62:1157–70. doi: 10.1128/mmbr.62.4.1157-1170.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bouskra D, et al. Lymphoid tissue genesis induced by commensals through NOD1 regulates intestinal homeostasis. Nature. 2008;456:507–10. doi: 10.1038/nature07450. [This study demonstrates that peptidoglycan from gram negative bacteria induces the generation of ILF's through the recongition of NOD1. In the absence of ILF formation, dramatic changes in the composition of the microbiota occur.] [DOI] [PubMed] [Google Scholar]
- 13.Abrams GD, Bauer H, Sprinz H. Influence of the normal flora on mucosal morphology and cellular renewal in the ileum. A comparison of germ-free and conventional mice. Lab Invest. 1963;12:355–64. [PubMed] [Google Scholar]
- 14.Bry L, Falk PG, Midtvedt T, Gordon JI. A model of host-microbial interactions in an open mammalian ecosystem. Science. 1996;273:1380–3. doi: 10.1126/science.273.5280.1380. [DOI] [PubMed] [Google Scholar]
- 15.Cash HL, Whitham CV, Behrendt CL, Hooper LV. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science. 2006;313:1126–30. doi: 10.1126/science.1127119. [These authors demonstrate that microbial colonization of germfree mice induce RegIIIγ, a secreted C-type lectin. They further show that this protein has antimicrobial activity by binding to peptidoglycan, suggesting that microbial species actively shape the intestinal environment to their advantage.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sonnenburg JL, Chen CT, Gordon JI. Genomic and metabolic studies of the impact of probiotics on a model gut symbiont and host. PLoS Biol. 2006;4:e413. doi: 10.1371/journal.pbio.0040413. [Using B.thetaiotaomicron (a symbiont) co-colonization with Bifidobacterium longum,(a probiotic) in germfree mice, this study demonstrates that B.longum can increase the diversity of polysaccarides that can be degraded by B.thetaiotaomicron, Thus, demonstrating another layer of complexity in host microbial interactions; that intestinal bacterial species can impact the function of one another.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sprinz H, et al. The response of the germfree guinea pig to oral bacterial challenge with Escherichia coli and Shigella flexneri. Am J Pathol. 1961;39:681–95. [PMC free article] [PubMed] [Google Scholar]
- 18.Maier BR, Hentges DJ. Experimental Shigella infections in laboratory animals. I. Antagonism by human normal flora components in gnotobiotic mice. Infect Immun. 1972;6:168–73. doi: 10.1128/iai.6.2.168-173.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zachar Z, Savage DC. Microbial interference and colonization of the murine gastrointestinal tract by Listeria monocytogenes. Infect Immun. 1979;23:168–74. doi: 10.1128/iai.23.1.168-174.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Inagaki H, Suzuki T, Nomoto K, Yoshikai Y. Increased susceptibility to primary infection with Listeria monocytogenes in germfree mice may be due to lack of accumulation of L-selectin+ CD44+ T cells in sites of inflammation. Infect Immun. 1996;64:3280–7. doi: 10.1128/iai.64.8.3280-3287.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nardi RM, Silva ME, Vieira EC, Bambirra EA, Nicoli JR. Intragastric infection of germfree and conventional mice with Salmonella typhimurium. Braz J Med Biol Res. 1989;22:1389–92. [PubMed] [Google Scholar]
- 22.Stecher B, et al. Salmonella enterica Serovar Typhimurium Exploits Inflammation to Compete with the Intestinal Microbiota. PLoS Biol. 2007;5:e244. doi: 10.1371/journal.pbio.0050244. [These authors demonstrate that that an intestinal pathogen, Salmonella enterica, utilizes inflammation in order to perturb the commensal microbial population to induce disease.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nardi RM, et al. Bacteriological and immunological aspects of conventional and germfree mice infected with Salmonella typhimurium. Rev Latinoam Microbiol. 1991;33:239–43. [PubMed] [Google Scholar]
- 24.Podolsky DK. The current future understanding of inflammatory bowel disease. Best Pract Res Clin Gastroenterol. 2002;16:933–43. doi: 10.1053/bega.2002.0354. [DOI] [PubMed] [Google Scholar]
- 25.Shanahan F. Crohn's disease. Lancet. 2002;359:62–9. doi: 10.1016/S0140-6736(02)07284-7. [DOI] [PubMed] [Google Scholar]
- 26.Targan SR, Karp LC. Defects in mucosal immunity leading to ulcerative colitis. Immunol Rev. 2005;206:296–305. doi: 10.1111/j.0105-2896.2005.00286.x. [DOI] [PubMed] [Google Scholar]
- 27.Bouma G, Strober W. The immunological and genetic basis of inflammatory bowel disease. Nat Rev Immunol. 2003;3:521–33. doi: 10.1038/nri1132. [DOI] [PubMed] [Google Scholar]
- 28.Kullberg MC, et al. IL-23 plays a key role in Helicobacter hepaticus-induced T cell-dependent colitis. J Exp Med. 2006;203:2485–94. doi: 10.1084/jem.20061082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hue S, et al. Interleukin-23 drives innate and T cell-mediated intestinal inflammation. J Exp Med. 2006;203:2473–83. doi: 10.1084/jem.20061099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Duerr RH, et al. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science. 2006;314:1461–3. doi: 10.1126/science.1135245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmechel S, et al. Linking genetic susceptibility to Crohn's disease with Th17 cell function: IL-22 serum levels are increased in Crohn's disease and correlate with disease activity and IL23R genotype status. Inflamm Bowel Dis. 2008;14:204–12. doi: 10.1002/ibd.20315. [DOI] [PubMed] [Google Scholar]
- 32.Kobayashi T, et al. IL23 differentially regulates the Th1/Th17 balance in ulcerative colitis and Crohn's disease. Gut. 2008;57:1682–9. doi: 10.1136/gut.2007.135053. [DOI] [PubMed] [Google Scholar]
- 33.Sartor RB. Microbial influences in inflammatory bowel diseases. Gastroenterology. 2008;134:577–94. doi: 10.1053/j.gastro.2007.11.059. [DOI] [PubMed] [Google Scholar]
- 34.Fontenot JD, Rudensky AY. A well adapted regulatory contrivance: regulatory T cell development and the forkhead family transcription factor Foxp3. Nat Immunol. 2005;6:331–7. doi: 10.1038/ni1179. [DOI] [PubMed] [Google Scholar]
- 35.Vignali DA, Collison LW, Workman CJ. How regulatory T cells work. Nat Rev Immunol. 2008;8:523–32. doi: 10.1038/nri2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Coombes JL, et al. A functionally specialized population of mucosal CD103+ DCs induces Foxp3+ regulatory T cells via a TGF-beta and retinoic acid-dependent mechanism. J Exp Med. 2007;204:1757–64. doi: 10.1084/jem.20070590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Powrie F, Maloy KJ. Immunology. Regulating the regulators. Science. 2003;299:1030–1. doi: 10.1126/science.1082031. [DOI] [PubMed] [Google Scholar]
- 38.Collison LW, et al. The inhibitory cytokine IL-35 contributes to regulatory T-cell function. Nature. 2007;450:566–9. doi: 10.1038/nature06306. [DOI] [PubMed] [Google Scholar]
- 39.Ivanov II, et al. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe. 2008;4:337–49. doi: 10.1016/j.chom.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Atarashi K, et al. ATP drives lamina propria T(H)17 cell differentiation. Nature. 2008;455:808–12. doi: 10.1038/nature07240. [DOI] [PubMed] [Google Scholar]
- 41.Hall JA, et al. Commensal DNA limits regulatory T cell conversion and is a natural adjuvant of intestinal immune responses. Immunity. 2008;29:637–49. doi: 10.1016/j.immuni.2008.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004;118:229–41. doi: 10.1016/j.cell.2004.07.002. [One of the first studies to suggest that recognition of TLR ligands of commensal bacteria by the host is important in the maintainence of intestinal homoestasis.] [DOI] [PubMed] [Google Scholar]
- 43.Ostman S, Rask C, Wold AE, Hultkrantz S, Telemo E. Impaired regulatory T cell function in germ-free mice. Eur J Immunol. 2006;36:2336–46. doi: 10.1002/eji.200535244. [DOI] [PubMed] [Google Scholar]
- 44.Ishikawa H, et al. Effect of intestinal microbiota on the induction of regulatory CD25+ CD4+ T cells. Clin Exp Immunol. 2008;153:127–35. doi: 10.1111/j.1365-2249.2008.03668.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Strauch UG, et al. Influence of intestinal bacteria on induction of regulatory T cells: lessons from a transfer model of colitis. Gut. 2005;54:1546–52. doi: 10.1136/gut.2004.059451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zaph C, et al. Commensal-dependent expression of IL-25 regulates the IL-23-IL-17 axis in the intestine. J Exp Med. 2008;205:2191–8. doi: 10.1084/jem.20080720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.De Winter H, Cheroutre H, Kronenberg M. Mucosal immunity and inflammation. II. The yin and yang of T cells in intestinal inflammation: pathogenic and protective roles in a mouse colitis model. Am J Physiol. 1999;276:G1317–21. doi: 10.1152/ajpgi.1999.276.6.G1317. [DOI] [PubMed] [Google Scholar]
- 48.Simpson SJ, de Jong YP, Comiskey M, Terhorst C. Pathways of T cell pathology in models of chronic intestinal inflammation. Int Rev Immunol. 2000;19:1–37. doi: 10.3109/08830180009048387. [DOI] [PubMed] [Google Scholar]
- 49.Elson CO, et al. Monoclonal anti-interleukin 23 reverses active colitis in a T cell-mediated model in mice. Gastroenterology. 2007;132:2359–70. doi: 10.1053/j.gastro.2007.03.104. [DOI] [PubMed] [Google Scholar]
- 50.Sartor RB. The influence of normal microbial flora on the development of chronic mucosal inflammation. Res Immunol. 1997;148:567–76. doi: 10.1016/s0923-2494(98)80151-x. [DOI] [PubMed] [Google Scholar]
- 51.Macpherson A, Khoo UY, Forgacs I, Philpott-Howard J, Bjarnason I. Mucosal antibodies in inflammatory bowel disease are directed against intestinal bacteria. Gut. 1996;38:365–75. doi: 10.1136/gut.38.3.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Elson CO. Commensal bacteria as targets in Crohn's disease. Gastroenterology. 2000;119:254–7. doi: 10.1053/gast.2000.9159. [DOI] [PubMed] [Google Scholar]
- 53.Tannock GW. Exploring the relationships between intestinal microflora and inflammatory conditions of the human bowel and spine. Antonie Van Leeuwenhoek. 2002;81:529–35. doi: 10.1023/a:1020517603993. [DOI] [PubMed] [Google Scholar]
- 54.Kent TH, Summers RW, DenBesten L, Swaner JC, Hrouda M. Effect of antibiotics on bacterial flora of rats with intestinal blind loops. Proc Soc Exp Biol Med. 1969;132:63–7. doi: 10.3181/00379727-132-34149. [DOI] [PubMed] [Google Scholar]
- 55.Videla S, et al. Role of intestinal microflora in chronic inflammation and ulceration of the rat colon. Gut. 1994;35:1090–7. doi: 10.1136/gut.35.8.1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Taurog JD, et al. The germfree state prevents development of gut and joint inflammatory disease in HLA-B27 transgenic rats. J Exp Med. 1994;180:2359–64. doi: 10.1084/jem.180.6.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rath HC. Role of commensal bacteria in chronic experimental colitis: lessons from the HLA-B27 transgenic rat. Pathobiology. 2002;70:131–8. doi: 10.1159/000068144. [DOI] [PubMed] [Google Scholar]
- 58.Sellon RK, et al. Resident enteric bacteria are necessary for development of spontaneous colitis and immune system activation in interleukin-10-deficient mice. Infect Immun. 1998;66:5224–31. doi: 10.1128/iai.66.11.5224-5231.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cahill RJ, et al. Inflammatory bowel disease: an immunity-mediated condition triggered by bacterial infection with Helicobacter hepaticus. Infect Immun. 1997;65:3126–31. doi: 10.1128/iai.65.8.3126-3131.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kullberg MC, et al. Induction of colitis by a CD4+ T cell clone specific for a bacterial epitope. Proc Natl Acad Sci U S A. 2003;100:15830–5. doi: 10.1073/pnas.2534546100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Barnich N, et al. CEACAM6 acts as a receptor for adherent-invasive E. coli, supporting ileal mucosa colonization in Crohn disease. J Clin Invest. 2007;117:1566–74. doi: 10.1172/JCI30504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hampe J, et al. Association between insertion mutation in NOD2 gene and Crohn's disease in German and British populations. Lancet. 2001;357:1925–8. doi: 10.1016/S0140-6736(00)05063-7. [DOI] [PubMed] [Google Scholar]
- 63.Kim SC, Tonkonogy SL, Karrasch T, Jobin C, Sartor RB. Dual-association of gnotobiotic IL-10−/− mice with 2 nonpathogenic commensal bacteria induces aggressive pancolitis. Inflamm Bowel Dis. 2007;13:1457–66. doi: 10.1002/ibd.20246. [DOI] [PubMed] [Google Scholar]
- 64.O'Hara AM, Shanahan F. The gut flora as a forgotten organ. EMBO Rep. 2006;7:688–93. doi: 10.1038/sj.embor.7400731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ley RE, Knight R, Gordon JI. The human microbiome: eliminating the biomedical/environmental dichotomy in microbial ecology. Environ Microbiol. 2007;9:3–4. doi: 10.1111/j.1462-2920.2006.01222_3.x. [DOI] [PubMed] [Google Scholar]
- 66.Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453:620–5. doi: 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- 67.Glimcher LH. Trawling for treasure: tales of T-bet. Nat Immunol. 2007;8:448–50. doi: 10.1038/ni0507-448. [DOI] [PubMed] [Google Scholar]
- 68.Garrett WS, et al. Communicable ulcerative colitis induced by T-bet deficiency in the innate immune system. Cell. 2007;131:33–45. doi: 10.1016/j.cell.2007.08.017. [This study demonstrates that mice deficient in the transcription factor T-bet within the innate immune system develop spontaneous colitis. More importantly, transfer of the microbiota from these mice into wild-type recipients induces disease.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Turnbaugh PJ, et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–31. doi: 10.1038/nature05414. [These authors show that the microbiome from obese mice has an increased capacity for energy harvest. Transfer of the microbiota to non-obese mice increases their mean fat body weight, suggesting that a change in the microbiota can induce disease.] [DOI] [PubMed] [Google Scholar]
- 70.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444:1022–3. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 71.Pomp D, Nehrenberg D, Estrada-Smith D. Complex genetics of obesity in mouse models. Annu Rev Nutr. 2008;28:331–45. doi: 10.1146/annurev.nutr.27.061406.093552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lepage P, et al. Biodiversity of the mucosa-associated microbiota is stable along the distal digestive tract in healthy individuals and patients with IBD. Inflamm Bowel Dis. 2005;11:473–80. doi: 10.1097/01.mib.0000159662.62651.06. [DOI] [PubMed] [Google Scholar]
- 73.Scanlan PD, Shanahan F, O'Mahony C, Marchesi JR. Culture-independent analyses of temporal variation of the dominant fecal microbiota and targeted bacterial subgroups in Crohn's disease. J Clin Microbiol. 2006;44:3980–8. doi: 10.1128/JCM.00312-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Frank DN, et al. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci U S A. 2007;104:13780–5. doi: 10.1073/pnas.0706625104. [This study demonstrates that the intestinal microbial populations between human IBD and non-IBD patients differs significantly.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Turnbaugh PJ, et al. The human microbiome project. Nature. 2007;449:804–10. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sartor RB. Therapeutic manipulation of the enteric microflora in inflammatory bowel diseases: antibiotics, probiotics, and prebiotics. Gastroenterology. 2004;126:1620–33. doi: 10.1053/j.gastro.2004.03.024. [DOI] [PubMed] [Google Scholar]
- 77.Di Giacinto C, Marinaro M, Sanchez M, Strober W, Boirivant M. Probiotics ameliorate recurrent Th1-mediated murine colitis by inducing IL-10 and IL-10-dependent TGF-beta-bearing regulatory cells. J Immunol. 2005;174:3237–46. doi: 10.4049/jimmunol.174.6.3237. [DOI] [PubMed] [Google Scholar]
- 78.O'Mahony C, et al. Commensal-induced regulatory T cells mediate protection against pathogen-stimulated NF-kappaB activation. PLoS Pathog. 2008;4:e1000112. doi: 10.1371/journal.ppat.1000112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Foligne B, et al. Correlation between in vitro and in vivo immunomodulatory properties of lactic acid bacteria. World J Gastroenterol. 2007;13:236–43. doi: 10.3748/wjg.v13.i2.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sokol H, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci U S A. 2008;105:16731–6. doi: 10.1073/pnas.0804812105. [These authors identify that the bacterium F. prausnitzii is specifically reduced within the intestine of Crohns patients. They further demonstrate that this bacterium has an anti-inflammatory capacity and can protect animals from disease when orally administered, suggesting that symbiotic microbes may be directly mediating health.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 2005;122:107–18. doi: 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 82.Roncarolo MG, Battaglia M. Regulatory T-cell immunotherapy for tolerance to self antigens and alloantigens in humans. Nat Rev Immunol. 2007;7:585–98. doi: 10.1038/nri2138. [DOI] [PubMed] [Google Scholar]
- 83.Cong Y, et al. Generation of antigen-specific, Foxp3-expressing CD4+ regulatory T cells by inhibition of APC proteosome function. J Immunol. 2005;174:2787–95. doi: 10.4049/jimmunol.174.5.2787. [DOI] [PubMed] [Google Scholar]
- 84.Noverr MC, Huffnagle GB. Does the microbiota regulate immune responses outside the gut? Trends Microbiol. 2004;12:562–8. doi: 10.1016/j.tim.2004.10.008. [DOI] [PubMed] [Google Scholar]
- 85.Bjorksten B. The environmental influence on childhood asthma. Allergy. 1999;54(Suppl 49):17–23. doi: 10.1111/j.1398-9995.1999.tb04383.x. [DOI] [PubMed] [Google Scholar]
- 86.Penders J, et al. Gut microbiota composition and development of atopic manifestations in infancy: the KOALA Birth Cohort Study. Gut. 2007;56:661–7. doi: 10.1136/gut.2006.100164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kalliomaki M, Isolauri E. Pandemic of atopic diseases--a lack of microbial exposure in early infancy? Curr Drug Targets Infect Disord. 2002;2:193–9. doi: 10.2174/1568005023342452. [DOI] [PubMed] [Google Scholar]
- 88.Kalliomaki M, Isolauri E. Role of intestinal flora in the development of allergy. Curr Opin Allergy Clin Immunol. 2003;3:15–20. doi: 10.1097/00130832-200302000-00003. [DOI] [PubMed] [Google Scholar]
- 89.Sakaguchi S, et al. Foxp3+ CD25+ CD4+ natural regulatory T cells in dominant self-tolerance and autoimmune disease. Immunol Rev. 2006;212:8–27. doi: 10.1111/j.0105-2896.2006.00427.x. [DOI] [PubMed] [Google Scholar]
- 90.Zhou L, et al. TGF-beta-induced Foxp3 inhibits T(H)17 cell differentiation by antagonizing RORgammat function. Nature. 2008;453:236–40. doi: 10.1038/nature06878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Eckburg PB, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–8. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gill SR, et al. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312:1355–9. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Palmer C, Bik EM, Digiulio DB, Relman DA, Brown PO. Development of the Human Infant Intestinal Microbiota. PLoS Biol. 2007;5:e177. doi: 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Izcue A, et al. Interleukin-23 restrains regulatory T cell activity to drive T cell-dependent colitis. Immunity. 2008;28:559–70. doi: 10.1016/j.immuni.2008.02.019. [This report demonstrates that Foxp3 deficient T cells can induce colitis in IL-23 deficient recipients, suggesting that disease can result in the absence of regulation.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.O'Mahony SM, et al. Early life stress alters behavior, immunity, and microbiota in rats: implications for irritable bowel syndrome and psychiatric illnesses. Biol Psychiatry. 2009;65:263–7. doi: 10.1016/j.biopsych.2008.06.026. [DOI] [PubMed] [Google Scholar]
- 96.Moreau MC, Ducluzeau R, Guy-Grand D, Muller MC. Increase in the population of duodenal immunoglobulin A plasmocytes in axenic mice associated with different living or dead bacterial strains of intestinal origin. Infect Immun. 1978;21:532–9. doi: 10.1128/iai.21.2.532-539.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Suzuki K, et al. Aberrant expansion of segmented filamentous bacteria in IgA-deficient gut. Proc Natl Acad Sci U S A. 2004;101:1981–6. doi: 10.1073/pnas.0307317101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kroese FG, de Waard R, Bos NA. B-1 cells and their reactivity with the murine intestinal microflora. Semin Immunol. 1996;8:11–8. doi: 10.1006/smim.1996.0003. [DOI] [PubMed] [Google Scholar]
- 99.Macpherson AJ, Uhr T. Induction of protective IgA by intestinal dendritic cells carrying commensal bacteria. Science. 2004;303:1662–5. doi: 10.1126/science.1091334. [DOI] [PubMed] [Google Scholar]
- 100.Macpherson AJ, Geuking MB, McCoy KD. Immune responses that adapt the intestinal mucosa to commensal intestinal bacteria. Immunology. 2005;115:153–62. doi: 10.1111/j.1365-2567.2005.02159.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.He B, et al. Intestinal bacteria trigger T cell-independent immunoglobulin A(2) class switching by inducing epithelial-cell secretion of the cytokine APRIL. Immunity. 2007;26:812–26. doi: 10.1016/j.immuni.2007.04.014. [DOI] [PubMed] [Google Scholar]
- 102.Cerutti A. The regulation of IgA class switching. Nat Rev Immunol. 2008;8:421–34. doi: 10.1038/nri2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Tezuka H, et al. Regulation of IgA production by naturally occurring TNF/iNOS-producing dendritic cells. Nature. 2007;448:929–33. doi: 10.1038/nature06033. [DOI] [PubMed] [Google Scholar]
- 104.Peterson DA, McNulty NP, Guruge JL, Gordon JI. IgA response to symbiotic bacteria as a mediator of gut homeostasis. Cell Host Microbe. 2007;2:328–39. doi: 10.1016/j.chom.2007.09.013. [DOI] [PubMed] [Google Scholar]